Journal Description
Genes
Genes
is a peer-reviewed, open access journal of genetics and genomics published monthly online by MDPI. The Spanish Society for Biochemistry and Molecular Biology (SEBBM) is affiliated with Genes and their members receive discounts on the article processing charges.
- Open Access— free for readers, with article processing charges (APC) paid by authors or their institutions.
- High Visibility: indexed within Scopus, SCIE (Web of Science), PubMed, MEDLINE, PMC, Embase, PubAg, and other databases.
- Journal Rank: JCR - Q2 (Genetics & Heredity) / CiteScore - Q2 (Genetics)
- Rapid Publication: manuscripts are peer-reviewed and a first decision is provided to authors approximately 16.5 days after submission; acceptance to publication is undertaken in 2.3 days (median values for papers published in this journal in the second half of 2023).
- Recognition of Reviewers: Reviewers who provide timely, thorough peer-review reports receive vouchers entitling them to a discount on the APC of their next publication in any MDPI journal, in appreciation of the work done.
Impact Factor:
3.5 (2022);
5-Year Impact Factor:
3.9 (2022)
Latest Articles
Genetic Mechanisms Driving Uterine Leiomyoma Pathobiology, Epidemiology, and Treatment
Genes 2024, 15(5), 558; https://doi.org/10.3390/genes15050558 (registering DOI) - 27 Apr 2024
Abstract
Uterine leiomyomas (ULs) are the most common benign tumor of the uterus. They can be associated with symptoms including abnormal uterine bleeding, pelvic pain, urinary frequency, and pregnancy complications. Despite the high prevalence of UL, its underlying pathophysiology mechanisms have historically been poorly
[...] Read more.
Uterine leiomyomas (ULs) are the most common benign tumor of the uterus. They can be associated with symptoms including abnormal uterine bleeding, pelvic pain, urinary frequency, and pregnancy complications. Despite the high prevalence of UL, its underlying pathophysiology mechanisms have historically been poorly understood. Several mechanisms of pathogenesis have been suggested, implicating various genes, growth factors, cytokines, chemokines, and microRNA aberrations. The purpose of this study is to summarize the current research on the relationship of genetics with UL. Specifically, we performed a literature review of published studies to identify how genetic aberrations drive pathophysiology, epidemiology, and therapeutic approaches of UL. With regards to pathophysiology, research has identified MED12 mutations, HMGA2 overexpression, fumarate hydratase deficiency, and cytogenetic abnormalities as contributors to the development of UL. Additionally, epigenetic modifications, such as histone acetylation and DNA methylation, have been identified as contributing to UL tumorigenesis. Specifically, UL stem cells have been found to contain a unique DNA methylation pattern compared to more differentiated UL cells, suggesting that DNA methylation has a role in tumorigenesis. On a population level, genome-wide association studies (GWASs) and epidemiologic analyses have identified 23 genetic loci associated with younger age at menarche and UL growth. Additionally, various GWASs have investigated genetic loci as potential drivers of racial disparities in UL incidence. For example, decreased expression of Cytohesin 4 in African Americans has been associated with increased UL risk. Recent studies have investigated various therapeutic options, including ten-eleven translocation proteins mediating DNA methylation, adenovirus vectors for drug delivery, and “suicide gene therapy” to induce apoptosis. Overall, improved understanding of the genetic and epigenetic drivers of UL on an individual and population level can propel the discovery of novel therapeutic options.
Full article
(This article belongs to the Special Issue Genetics and Genomics of Female Reproduction)
►
Show Figures
Open AccessArticle
The Relationship between miR-5682 and Nutritional Status of Radiotherapy-Treated Male Laryngeal Cancer Patients
by
Marcin Mazurek, Anna Brzozowska, Mirosław Maziarz, Teresa Małecka-Massalska and Tomasz Powrózek
Genes 2024, 15(5), 556; https://doi.org/10.3390/genes15050556 (registering DOI) - 27 Apr 2024
Abstract
Background: Nutritional deficiencies are frequently observed in patients with head and neck cancer (HNC) undergoing radiation therapy. microRNAs (miRNAs) were found to play an important role in the development of metabolic disorders throughout regulation of genes involved in inflammatory responses. This study aimed
[...] Read more.
Background: Nutritional deficiencies are frequently observed in patients with head and neck cancer (HNC) undergoing radiation therapy. microRNAs (miRNAs) were found to play an important role in the development of metabolic disorders throughout regulation of genes involved in inflammatory responses. This study aimed to explore the correlation between pre-treatment miR-5682 expression and parameters reflecting nutritional deficits in laryngeal cancer (LC) patients subjected to radiotherapy (RT). Methods: Expression of miR-5682 was analyzed in plasma samples of 56 male LC individuals. Nutritional status of LC patients was assessed using anthropometric and laboratory parameters, bioelectrical impedance analysis (BIA) and clinical questionnaires. Results: A high expression of miR-5682 was associated with significantly lower values of BMI, fat mass, fat-free mass and plasma albumin at selected periods of RT course. miR-5682 allowed us to distinguish between patients classified with both SGA-C and low albumin level from other LC patients with 100% sensitivity and 69.6% specificity (AUC = 0.820; p < 0.0001). Higher expression of studied miRNA was significantly associated with shorter median overall survival (OS) in LC patients (HR = 2.26; p = 0.008). Conclusions: analysis of miR-5682 expression demonstrates a potential clinical utility in selection of LC patients suffering from nutritional deficiencies developing as a consequence of RT-based therapy.
Full article
(This article belongs to the Special Issue Non-coding RNAs in Human Health and Disease)
Open AccessArticle
GhCLCc-1, a Chloride Channel Gene from Upland Cotton, Positively Regulates Salt Tolerance by Modulating the Accumulation of Chloride Ions
by
Wenhao Li, Siqi Gao, Yinghao Zhao, Yuchen Wu, Xiaona Li, Jianing Li, Wei Zhu, Zongbin Ma and Wei Liu
Genes 2024, 15(5), 555; https://doi.org/10.3390/genes15050555 (registering DOI) - 26 Apr 2024
Abstract
The ionic toxicity induced by salinization has adverse effects on the growth and development of crops. However, researches on ionic toxicity and salt tolerance in plants have focused primarily on cations such as sodium ions (Na+), with very limited studies on
[...] Read more.
The ionic toxicity induced by salinization has adverse effects on the growth and development of crops. However, researches on ionic toxicity and salt tolerance in plants have focused primarily on cations such as sodium ions (Na+), with very limited studies on chloride ions (Cl−). Here, we cloned the homologous genes of Arabidopsis thaliana AtCLCc, GhCLCc-1A/D, from upland cotton (Gossypium hirsutum), which were significantly induced by NaCl or KCl treatments. Subcellular localization showed that GhCLCc-1A/D were both localized to the tonoplast. Complementation of Arabidopsis atclcc mutant with GhCLCc-1 rescued its salt-sensitive phenotype. In addition, the silencing of the GhCLCc-1 gene led to an increased accumulation of Cl− in the roots, stems, and leaves of cotton seedlings under salt treatments, resulting in compromised salt tolerance. And ectopic expression of the GhCLCc-1 gene in Arabidopsis reduced the accumulation of Cl− in transgenic lines under salt treatments, thereby enhancing salt tolerance. These findings elucidate that GhCLCc-1 positively regulates salt tolerance by modulating Cl− accumulation and could be a potential target gene for improving salt tolerance in plants.
Full article
(This article belongs to the Special Issue Cotton Genes, Genetics, and Genomics)
Open AccessArticle
Exploring the Role of the MUTYH Gene in Breast, Ovarian and Endometrial Cancer
by
Carla Lintas, Benedetta Canalis, Alessia Azzarà, Giovanna Sabarese, Giuseppe Perrone and Fiorella Gurrieri
Genes 2024, 15(5), 554; https://doi.org/10.3390/genes15050554 - 26 Apr 2024
Abstract
Background: MUTYH germline monoallelic variants have been detected in a number of patients affected by breast/ovarian cancer or endometrial cancer, suggesting a potential susceptibility role, though their significance remains elusive since the disease mechanism is normally recessive. Hence, the aim of this research
[...] Read more.
Background: MUTYH germline monoallelic variants have been detected in a number of patients affected by breast/ovarian cancer or endometrial cancer, suggesting a potential susceptibility role, though their significance remains elusive since the disease mechanism is normally recessive. Hence, the aim of this research was to explore the hypothesis that a second hit could have arisen in the other allele in the tumor tissue. Methods: we used Sanger sequencing and immunohistochemistry to search for a second MUTYH variant in the tumoral DNA and to assess protein expression, respectively. Results: we detected one variant of unknown significance, one variant with conflicting interpretation of pathogenicity and three benign/likely benign variants; the MUTYH protein was not detected in the tumor tissue of half of the patients, and in others, its expression was reduced. Conclusions: our results fail to demonstrate that germinal monoallelic MUTYH variants increase cancer risk through a LOH (loss of heterozygosity) mechanism in the somatic tissue; however, the absence or partial loss of the MUTYH protein in many tumors suggests its dysregulation regardless of MUTYH genetic status.
Full article
(This article belongs to the Special Issue Genetics of Multifactorial Diseases)
Open AccessReview
Exploring the Role of Cell-Free Nucleic Acids and Peritoneal Dialysis: A Narrative Review
by
Niccolò Morisi, Grazia Maria Virzì, Marco Ferrarini, Gaetano Alfano, Monica Zanella, Claudio Ronco and Gabriele Donati
Genes 2024, 15(5), 553; https://doi.org/10.3390/genes15050553 - 26 Apr 2024
Abstract
Introduction: Cell-free nucleic acids (cf-NAs) represent a promising biomarker of various pathological and physiological conditions. Since its discovery in 1948, cf-NAs gained prognostic value in oncology, immunology, and other relevant fields. In peritoneal dialysis (PD), blood purification is performed by exposing the
[...] Read more.
Introduction: Cell-free nucleic acids (cf-NAs) represent a promising biomarker of various pathological and physiological conditions. Since its discovery in 1948, cf-NAs gained prognostic value in oncology, immunology, and other relevant fields. In peritoneal dialysis (PD), blood purification is performed by exposing the peritoneal membrane. Relevant sections: Complications of PD such as acute peritonitis and peritoneal membrane aging are often critical in PD patient management. In this review, we focused on bacterial DNA, cell-free DNA, mitochondrial DNA (mtDNA), microRNA (miRNA), and their potential uses as biomarkers for monitoring PD and its complications. For instance, the isolation of bacterial DNA in early acute peritonitis allows bacterial identification and subsequent therapy implementation. Cell-free DNA in peritoneal dialysis effluent (PDE) represents a marker of stress of the peritoneal membrane in both acute and chronic PD complications. Moreover, miRNA are promising hallmarks of peritoneal membrane remodeling and aging, even before its manifestation. In this scenario, with multiple cytokines involved, mtDNA could be considered equally meaningful to determine tissue inflammation. Conclusions: This review explores the relevance of cf-NAs in PD, demonstrating its promising role for both diagnosis and treatment. Further studies are necessary to implement the use of cf-NAs in PD clinical practice.
Full article
(This article belongs to the Section Molecular Genetics and Genomics)
Open AccessArticle
Novel Genome-Engineered H Alleles Differentially Affect Lateral Inhibition and Cell Dichotomy Processes during Bristle Organ Development
by
Tanja C. Mönch, Thomas K. Smylla, Franziska Brändle, Anette Preiss and Anja C. Nagel
Genes 2024, 15(5), 552; https://doi.org/10.3390/genes15050552 - 26 Apr 2024
Abstract
Hairless (H) encodes the major antagonist in the Notch signaling pathway, which governs cellular differentiation of various tissues in Drosophila. By binding to the Notch signal transducer Suppressor of Hairless (Su(H)), H assembles repressor complexes onto Notch target genes. Using genome engineering,
[...] Read more.
Hairless (H) encodes the major antagonist in the Notch signaling pathway, which governs cellular differentiation of various tissues in Drosophila. By binding to the Notch signal transducer Suppressor of Hairless (Su(H)), H assembles repressor complexes onto Notch target genes. Using genome engineering, three new H alleles, HFA, HLLAAand HWA were generated and a phenotypic series was established by several parameters, reflecting the residual H-Su(H) binding capacity. Occasionally, homozygous HWA flies develop to adulthood. They were compared with the likewise semi-viable HNN allele affecting H-Su(H) nuclear entry. The H homozygotes were short-lived, sterile and flightless, yet showed largely normal expression of several mitochondrial genes. Typical for H mutants, both HWA and HNN homozygous alleles displayed strong defects in wing venation and mechano-sensory bristle development. Strikingly, however, HWA displayed only a loss of bristles, whereas bristle organs of HNN flies showed a complete shaft-to-socket transformation. Apparently, the impact of HWA is restricted to lateral inhibition, whereas that of HNN also affects the respective cell type specification. Notably, reduction in Su(H) gene dosage only suppressed the HNN bristle phenotype, but amplified that of HWA. We interpret these differences as to the role of H regarding Su(H) stability and availability.
Full article
(This article belongs to the Special Issue Gene Editing in Drosophila to Study Gene Function and Developmental Processes)
Open AccessArticle
Whole-Genome Resequencing Revealed Selective Signatures for Growth Traits in Hu and Gangba Sheep
by
Peifu Yang, Mingyu Shang, Jingjing Bao, Tianyi Liu, Jinke Xiong, Jupeng Huang, Jinghua Sun and Li Zhang
Genes 2024, 15(5), 551; https://doi.org/10.3390/genes15050551 - 26 Apr 2024
Abstract
A genomic study was conducted to uncover the selection signatures in sheep that show extremely significant differences in growth traits under the same breed, age in months, nutrition level, and management practices. Hu sheep from Gansu Province and Gangba sheep from the Tibet
[...] Read more.
A genomic study was conducted to uncover the selection signatures in sheep that show extremely significant differences in growth traits under the same breed, age in months, nutrition level, and management practices. Hu sheep from Gansu Province and Gangba sheep from the Tibet Autonomous Region in China were selected. We collected whole-genome data from 40 sheep individuals (24 Hu sheep and 16 Gangba sheep), through whole-genome sequencing. Selection signals were analyzed using parameters such as FST, π ratio, and Tajima’s D. We have identified several candidate genes that have undergone strong selection, particularly those associated with growth traits. Specifically, five growth-related genes were identified in both the Hu sheep group (HDAC1, MYH7B, LCK, ACVR1, GNAI2) and the Gangba sheep group (RBBP8, ACSL3, FBXW11, PLAT, CRB1). Additionally, in a genomic region strongly selected in both the Hu and Gangba sheep groups (Chr 22: 51,425,001-51,500,000), the growth-associated gene CYP2E1 was identified, further highlighting the genetic factors influencing growth characteristics in these breeds. This study analyzes the genetic basis for significant differences in sheep phenotypes, identifies candidate genes related to sheep growth traits, lays the foundation for molecular genetic breeding in sheep, and accelerates the genetic improvement in livestock.
Full article
(This article belongs to the Section Animal Genetics and Genomics)
►▼
Show Figures
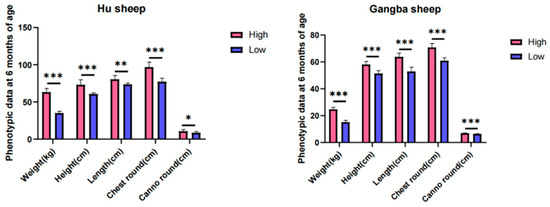
Figure 1
Open AccessArticle
Complete Chloroplast Genome Sequence Structure and Phylogenetic Analysis of Kohlrabi (Brassica oleracea var. gongylodes L.)
by
Mengliang Zhao, Yanxun Wu and Yanjing Ren
Genes 2024, 15(5), 550; https://doi.org/10.3390/genes15050550 - 26 Apr 2024
Abstract
Kohlrabi is an important swollen-stem cabbage variety belonging to the Brassicaceae family. However, few complete chloroplast genome sequences of this genus have been reported. Here, a complete chloroplast genome with a quadripartite cycle of 153,364 bp was obtained. A total of 132 genes
[...] Read more.
Kohlrabi is an important swollen-stem cabbage variety belonging to the Brassicaceae family. However, few complete chloroplast genome sequences of this genus have been reported. Here, a complete chloroplast genome with a quadripartite cycle of 153,364 bp was obtained. A total of 132 genes were identified, including 87 protein-coding genes, 37 transfer RNA genes and eight ribosomal RNA genes. The base composition analysis showed that the overall GC content was 36.36% of the complete chloroplast genome sequence. Relative synonymous codon usage frequency (RSCU) analysis showed that most codons with values greater than 1 ended with A or U, while most codons with values less than 1 ended with C or G. Thirty-five scattered repeats were identified and most of them were distributed in the large single-copy (LSC) region. A total of 290 simple sequence repeats (SSRs) were found and 188 of them were distributed in the LSC region. Phylogenetic relationship analysis showed that five Brassica oleracea subspecies were clustered into one group and the kohlrabi chloroplast genome was closely related to that of B. oleracea var. botrytis. Our results provide a basis for understanding chloroplast-dependent metabolic studies and provide new insight for understanding the polyploidization of Brassicaceae species.
Full article
(This article belongs to the Special Issue Plant Plastid Genome and Phylogenetics)
►▼
Show Figures

Figure 1
Open AccessArticle
A Bidirectional Non-Coding RNA Promoter Mediates Long-Range Gene Expression Regulation
by
Carlos Alberto Peralta-Alvarez, Hober Nelson Núñez-Martínez, Ángel Josué Cerecedo-Castillo, Augusto César Poot-Hernández, Gustavo Tapia-Urzúa, Sylvia Garza-Manero, Georgina Guerrero and Félix Recillas-Targa
Genes 2024, 15(5), 549; https://doi.org/10.3390/genes15050549 - 25 Apr 2024
Abstract
Recent evidence suggests that human gene promoters display gene expression regulatory mechanisms beyond the typical single gene local transcription modulation. In mammalian genomes, genes with an associated bidirectional promoter are abundant; bidirectional promoter architecture serves as a regulatory hub for a gene pair
[...] Read more.
Recent evidence suggests that human gene promoters display gene expression regulatory mechanisms beyond the typical single gene local transcription modulation. In mammalian genomes, genes with an associated bidirectional promoter are abundant; bidirectional promoter architecture serves as a regulatory hub for a gene pair expression. However, it has been suggested that its contribution to transcriptional regulation might exceed local transcription initiation modulation. Despite their abundance, the functional consequences of bidirectional promoter architecture remain largely unexplored. This work studies the long-range gene expression regulatory role of a long non-coding RNA gene promoter using chromosome conformation capture methods. We found that this particular bidirectional promoter contributes to distal gene expression regulation in a target-specific manner by establishing promoter–promoter interactions. In particular, we validated that the promoter–promoter interactions of this regulatory element with the promoter of distal gene BBX contribute to modulating the transcription rate of this gene; removing the bidirectional promoter from its genomic context leads to a rearrangement of BBX promoter–enhancer interactions and to increased gene expression. Moreover, long-range regulatory functionality is not directly dependent on its associated non-coding gene pair expression levels.
Full article
(This article belongs to the Section Molecular Genetics and Genomics)
Open AccessArticle
Non-Specific Epileptic Activity, EEG, and Brain Imaging in Loss of Function Variants in SATB1: A New Case Report and Review of the Literature
by
Flavia Privitera, Stefano Pagano, Camilla Meossi, Roberta Battini, Emanuele Bartolini, Domenico Montanaro and Filippo Maria Santorelli
Genes 2024, 15(5), 548; https://doi.org/10.3390/genes15050548 - 25 Apr 2024
Abstract
SATB1 (MIM #602075) is a relatively new gene reported only in recent years in association with neurodevelopmental disorders characterized by variable facial dysmorphisms, global developmental delay, poor or absent speech, altered electroencephalogram (EEG), and brain abnormalities on imaging. To date about thirty variants
[...] Read more.
SATB1 (MIM #602075) is a relatively new gene reported only in recent years in association with neurodevelopmental disorders characterized by variable facial dysmorphisms, global developmental delay, poor or absent speech, altered electroencephalogram (EEG), and brain abnormalities on imaging. To date about thirty variants in forty-four patients/children have been described, with a heterogeneous spectrum of clinical manifestations. In the present study, we describe a new patient affected by mild intellectual disability, speech disorder, and non-specific abnormalities on EEG and neuroimaging. Family studies identified a new de novo frameshift variant c.1818delG (p.(Gln606Hisfs*101)) in SATB1. To better define genotype–phenotype associations in the different types of reported SATB1 variants, we reviewed clinical data from our patient and from the literature and compared manifestations (epileptic activity, EEG abnormalities and abnormal brain imaging) due to missense variants versus those attributable to loss-of-function/premature termination variants. Our analyses showed that the latter variants are associated with less severe, non-specific clinical features when compared with the more severe phenotypes due to missense variants. These findings provide new insights into SATB1-related disorders.
Full article
(This article belongs to the Section Genetic Diagnosis)
Open AccessArticle
Statistical Genetic Approaches to Investigate Genotype-by-Environment Interaction: Review and Novel Extension of Models
by
Vincent P. Diego, Eron G. Manusov, Marcio Almeida, Sandra Laston, David Ortiz, John Blangero and Sarah Williams-Blangero
Genes 2024, 15(5), 547; https://doi.org/10.3390/genes15050547 - 25 Apr 2024
Abstract
Statistical genetic models of genotype-by-environment (G×E) interaction can be divided into two general classes, one on G×E interaction in response to dichotomous environments (e.g., sex, disease-affection status, or presence/absence of an exposure) and the other in response to continuous environments (e.g., physical activity,
[...] Read more.
Statistical genetic models of genotype-by-environment (G×E) interaction can be divided into two general classes, one on G×E interaction in response to dichotomous environments (e.g., sex, disease-affection status, or presence/absence of an exposure) and the other in response to continuous environments (e.g., physical activity, nutritional measurements, or continuous socioeconomic measures). Here we develop a novel model to jointly account for dichotomous and continuous environments. We develop the model in terms of a joint genotype-by-sex (for the dichotomous environment) and genotype-by-social determinants of health (SDoH; for the continuous environment). Using this model, we show how a depression variable, as measured by the Beck Depression Inventory-II survey instrument, is not only underlain by genetic effects (as has been reported elsewhere) but is also significantly determined by joint G×Sex and G×SDoH interaction effects. This model has numerous applications leading to potentially transformative research on the genetic and environmental determinants underlying complex diseases.
Full article
(This article belongs to the Special Issue Statistical Genetics of Human Complex Traits)
►▼
Show Figures
Figure 1
Open AccessArticle
Complete Chloroplast Genome of Krascheninnikovia ewersmanniana: Comparative and Phylogenetic Analysis
by
Peng Wei, Youzheng Li, Mei Ke, Yurong Hou, Abudureyimu Aikebaier and Zinian Wu
Genes 2024, 15(5), 546; https://doi.org/10.3390/genes15050546 - 25 Apr 2024
Abstract
Krascheninnikovia ewersmanniana is a dominant desert shrub in Xinjiang, China, with high economic and ecological value. However, molecular systematics research on K. ewersmanniana is lacking. To resolve the genetic composition of K. ewersmanniana within Amaranthaceae and its systematic relationship with
[...] Read more.
Krascheninnikovia ewersmanniana is a dominant desert shrub in Xinjiang, China, with high economic and ecological value. However, molecular systematics research on K. ewersmanniana is lacking. To resolve the genetic composition of K. ewersmanniana within Amaranthaceae and its systematic relationship with related genera, we used a second-generation Illumina sequencing system to detect the chloroplast genome of K. ewersmanniana and analyze its assembly, annotation, and phylogenetics. Total length of the chloroplast genome of K. ewersmanniana reached 152,287 bp, with 84 protein-coding genes, 36 tRNAs, and eight rRNAs. Codon usage analysis showed the majority of codons ending with base A/U. Mononucleotide repeats were the most common (85.42%) of the four identified simple sequence repeats. A comparison with chloroplast genomes of six other Amaranthaceae species indicated contraction and expansion of the inverted repeat boundary region in K. ewersmanniana, with some genes (rps19, ndhF, ycf1) differing in length and distribution. Among the seven species, the variation in non-coding regions was greater. Phylogenetic analysis revealed Krascheninnikovia ceratoides, Dysphania ambrosioides, Dysphania pumilio, and Dysphania botrys to have a close monophyletic relationship. By sequencing the K. ewersmanniana chloroplast genome, this research resolves the relatedness among 35 Amaranthaceae species, providing molecular insights for germplasm utilization, and theoretical support for studying evolutionary relationships.
Full article
(This article belongs to the Special Issue Advances in Evolution of Plant Organelle Genome (Volume II))
Open AccessArticle
Integrated Analysis of Transcriptome and Metabolome Reveals Differential Responses to Alternaria brassicicola Infection in Cabbage (Brassica oleracea var. capitata)
by
Jinzhou Lei, Wei Zhang, Fangwei Yu, Meng Ni, Zhigang Liu, Cheng Wang, Jianbin Li, Jianghua Song and Shenyun Wang
Genes 2024, 15(5), 545; https://doi.org/10.3390/genes15050545 - 25 Apr 2024
Abstract
Black spot, caused by Alternaria brassicicola (Ab), poses a serious threat to crucifer production, and knowledge of how plants respond to Ab infection is essential for black spot management. In the current study, combined transcriptomic and metabolic analysis was employed to
[...] Read more.
Black spot, caused by Alternaria brassicicola (Ab), poses a serious threat to crucifer production, and knowledge of how plants respond to Ab infection is essential for black spot management. In the current study, combined transcriptomic and metabolic analysis was employed to investigate the response to Ab infection in two cabbage (Brassica oleracea var. capitata) genotypes, Bo257 (resistant to Ab) and Bo190 (susceptible to Ab). A total of 1100 and 7490 differentially expressed genes were identified in Bo257 (R_mock vs. R_Ab) and Bo190 (S_mock vs. S_Ab), respectively. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis revealed that “metabolic pathways”, “biosynthesis of secondary metabolites”, and “glucosinolate biosynthesis” were the top three enriched KEGG pathways in Bo257, while “metabolic pathways”, “biosynthesis of secondary metabolites”, and “carbon metabolism” were the top three enriched KEGG pathways in Bo190. Further analysis showed that genes involved in extracellular reactive oxygen species (ROS) production, jasmonic acid signaling pathway, and indolic glucosinolate biosynthesis pathway were differentially expressed in response to Ab infection. Notably, when infected with Ab, genes involved in extracellular ROS production were largely unchanged in Bo257, whereas most of these genes were upregulated in Bo190. Metabolic profiling revealed 24 and 56 differentially accumulated metabolites in Bo257 and Bo190, respectively, with the majority being primary metabolites. Further analysis revealed that dramatic accumulation of succinate was observed in Bo257 and Bo190, which may provide energy for resistance responses against Ab infection via the tricarboxylic acid cycle pathway. Collectively, this study provides comprehensive insights into the Ab–cabbage interactions and helps uncover targets for breeding Ab-resistant varieties in cabbage.
Full article
(This article belongs to the Special Issue Vegetable Genetic Breeding)
►▼
Show Figures
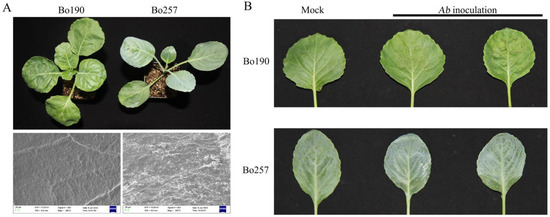
Figure 1
Open AccessArticle
Complete Chloroplast Genome of Alternanthera sessilis and Comparative Analysis with Its Congeneric Invasive Weed Alternanthera philoxeroides
by
Yuanxin Wang, Xueying Zhao, Qianhui Chen, Jun Yang, Jun Hu, Dong Jia and Ruiyan Ma
Genes 2024, 15(5), 544; https://doi.org/10.3390/genes15050544 - 25 Apr 2024
Abstract
Alternanthera sessilis is considered the closest relative to the invasive weed Alternanthera philoxeroides in China, making it an important native species for studying the invasive mechanisms and adaptations of A. philoxeroides. Chloroplasts play a crucial role in a plant’s environmental adaptation, with
[...] Read more.
Alternanthera sessilis is considered the closest relative to the invasive weed Alternanthera philoxeroides in China, making it an important native species for studying the invasive mechanisms and adaptations of A. philoxeroides. Chloroplasts play a crucial role in a plant’s environmental adaptation, with their genomes being pivotal in the evolution and adaptation of both invasive and related species. However, the chloroplast genome of A. sessilis has remained unknown until now. In this study, we sequenced and assembled the complete chloroplast genome of A. sessilis using high-throughput sequencing. The A. sessilis chloroplast genome is 151,935 base pairs long, comprising two inverted repeat regions, a large single copy region, and a small single copy region. This chloroplast genome contains 128 genes, including 8 rRNA-coding genes, 37 tRNA-coding genes, 4 pseudogenes, and 83 protein-coding genes. When compared to the chloroplast genome of the invasive weed A. philoxeroides and other Amaranthaceae species, we observed significant variations in the ccsA, ycf1, and ycf2 regions in the A. sessilis chloroplast genome. Moreover, two genes, ccsA and accD, were found to be undergoing rapid evolution due to positive selection pressure. The phylogenetic trees were constructed for the Amaranthaceae family, estimating the time of independent species formation between A. philoxeroides and A. sessilis to be approximately 3.5186–8.8242 million years ago. These findings provide a foundation for understanding the population variation within invasive species among the Alternanthera genus.
Full article
(This article belongs to the Section Plant Genetics and Genomics)
►▼
Show Figures
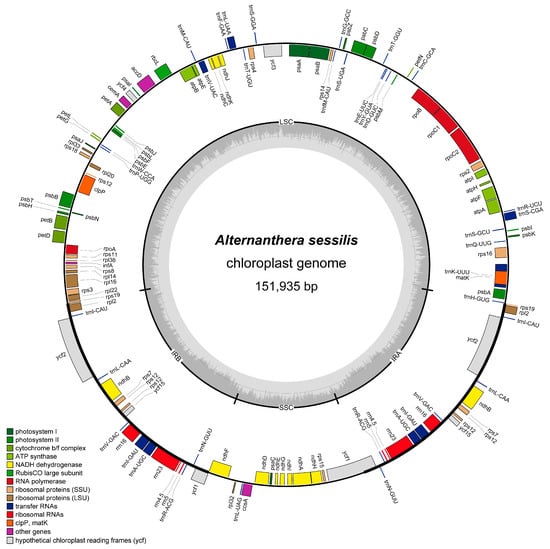
Figure 1
Open AccessArticle
Downregulation of Histone H3.3 Induces p53-Dependent Cellular Senescence in Human Diploid Fibroblasts
by
Yuki Yamamoto, Ryou-u Takahashi, Masaki Kinehara, Kimiyoshi Yano, Tatsuya Kuramoto, Akira Shimamoto and Hidetoshi Tahara
Genes 2024, 15(5), 543; https://doi.org/10.3390/genes15050543 - 25 Apr 2024
Abstract
Cellular senescence is an irreversible growth arrest that acts as a barrier to cancer initiation and progression. Histone alteration is one of the major events during replicative senescence. However, little is known about the function of H3.3 in cellular senescence. Here we found
[...] Read more.
Cellular senescence is an irreversible growth arrest that acts as a barrier to cancer initiation and progression. Histone alteration is one of the major events during replicative senescence. However, little is known about the function of H3.3 in cellular senescence. Here we found that the downregulation of H3.3 induced growth suppression with senescence-like phenotypes such as senescence-associated heterochromatin foci (SAHF) and β-galactosidase (SA-β-gal) activity. Furthermore, H3.3 depletion induced senescence-like phenotypes with the p53/p21-depedent pathway. In addition, we identified miR-22-3p, tumor suppressive miRNA, as an upstream regulator of the H3F3B (H3 histone, family 3B) gene which is the histone variant H3.3 and replaces conventional H3 in active genes. Therefore, our results reveal for the first time the molecular mechanisms for cellular senescence which are regulated by H3.3 abundance. Taken together, our studies suggest that H3.3 exerts functional roles in regulating cellular senescence and is a promising target for cancer therapy.
Full article
(This article belongs to the Section Human Genomics and Genetic Diseases)
►▼
Show Figures
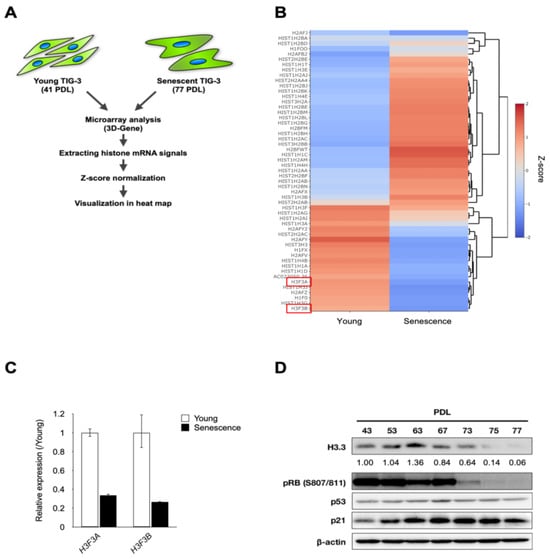
Figure 1
Open AccessArticle
Comparative Analysis of Virulence and Molecular Diversity of Puccinia striiformis f. sp. tritici Isolates Collected in 2016 and 2023 in the Western Region of China
by
Tesfay Gebrekirstos Gebremariam, Fengtao Wang, Ruiming Lin and Hongjie Li
Genes 2024, 15(5), 542; https://doi.org/10.3390/genes15050542 - 25 Apr 2024
Abstract
Puccinia striiformis f. sp. tritici (Pst) is adept at overcoming resistance in wheat cultivars, through variations in virulence in the western provinces of China. To apply disease management strategies, it is essential to understand the temporal and spatial dynamics of Pst
[...] Read more.
Puccinia striiformis f. sp. tritici (Pst) is adept at overcoming resistance in wheat cultivars, through variations in virulence in the western provinces of China. To apply disease management strategies, it is essential to understand the temporal and spatial dynamics of Pst populations. This study aimed to evaluate the virulence and molecular diversity of 84 old Pst isolates, in comparison to 59 newer ones. By using 19 Chinese wheat differentials, we identified 98 pathotypes, showing virulence complexity ranging from 0 to 16. Associations between 23 Yr gene pairs showed linkage disequilibrium and have the potential for gene pyramiding. The new Pst isolates had a higher number of polymorphic alleles (1.97), while the older isolates had a slightly higher number of effective alleles, Shannon’s information, and diversity. The Gansu Pst population had the highest diversity (uh = 0.35), while the Guizhou population was the least diverse. Analysis of molecular variance revealed that 94% of the observed variation occurred within Pst populations across the four provinces, while 6% was attributed to differences among populations. Overall, Pst populations displayed a higher pathotypic diversity of H > 2.5 and a genotypic diversity of 96%. This underscores the need to develop gene-pyramided cultivars to enhance the durability of resistance.
Full article
(This article belongs to the Special Issue Quality Gene Mining and Breeding of Wheat)
►▼
Show Figures
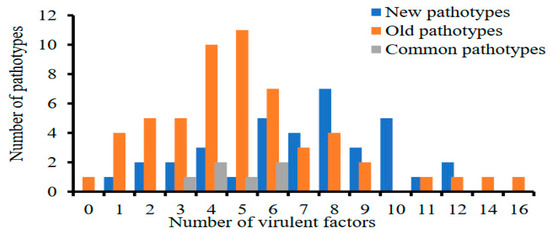
Figure 1
Open AccessArticle
The Satellite DNA PcH-Sat, Isolated and Characterized in the Limpet Patella caerulea (Mollusca, Gastropoda), Suggests the Origin from a Nin-SINE Transposable Element
by
Agnese Petraccioli, Nicola Maio, Rosa Carotenuto, Gaetano Odierna and Fabio Maria Guarino
Genes 2024, 15(5), 541; https://doi.org/10.3390/genes15050541 - 25 Apr 2024
Abstract
Satellite DNA (sat-DNA) was previously described as junk and selfish DNA in the cellular economy, without a clear functional role. However, during the last two decades, evidence has been accumulated about the roles of sat-DNA in different cellular functions and its probable involvement
[...] Read more.
Satellite DNA (sat-DNA) was previously described as junk and selfish DNA in the cellular economy, without a clear functional role. However, during the last two decades, evidence has been accumulated about the roles of sat-DNA in different cellular functions and its probable involvement in tumorigenesis and adaptation to environmental changes. In molluscs, studies on sat-DNAs have been performed mainly on bivalve species, especially those of economic interest. Conversely, in Gastropoda (which includes about 80% of the currently described molluscs species), studies on sat-DNA have been largely neglected. In this study, we isolated and characterized a sat-DNA, here named PcH-sat, in the limpet Patella caerulea using the restriction enzyme method, particularly HaeIII. Monomeric units of PcH-sat are 179 bp long, AT-rich (58.7%), and with an identity among monomers ranging from 91.6 to 99.8%. Southern blot showed that PcH-sat is conserved in P. depressa and P. ulyssiponensis, while a smeared signal of hybridization was present in the other three investigated limpets (P. ferruginea, P. rustica and P. vulgata). Dot blot showed that PcH-sat represents about 10% of the genome of P. caerulea, 5% of that of P. depressa, and 0.3% of that of P. ulyssiponensis. FISH showed that PcH-sat was mainly localized on pericentromeric regions of chromosome pairs 2 and 4–7 of P. caerulea (2n = 18). A database search showed that PcH-sat contains a large segment (of 118 bp) showing high identity with a homologous trait of the Nin-SINE transposable element (TE) of the patellogastropod Lottia gigantea, supporting the hypothesis that TEs are involved in the rising and tandemization processes of sat-DNAs.
Full article
(This article belongs to the Section Animal Genetics and Genomics)
►▼
Show Figures
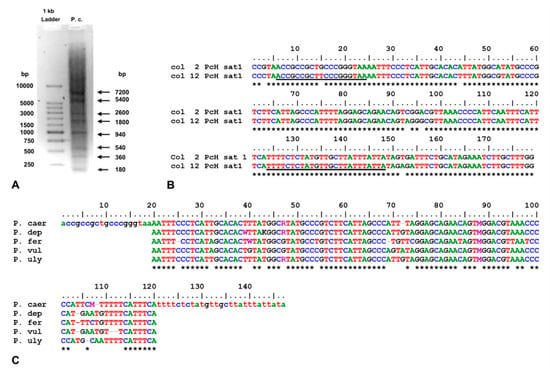
Figure 1
Open AccessArticle
Characterization of the Apoptotic and Antimicrobial Activities of Two Initiator Caspases of Sea Cucumber Apostichopus japonicus
by
Hanshuo Zhu, Zihao Yuan, Hang Xu and Li Sun
Genes 2024, 15(5), 540; https://doi.org/10.3390/genes15050540 - 25 Apr 2024
Abstract
Caspase (CASP) is a protease family that plays a vital role in apoptosis, development, and immune response. Herein, we reported the identification and characterization of two CASPs, AjCASPX1 and AjCASPX2, from the sea cucumber Apostichopus japonicus, an important aquaculture species. AjCASPX1/2 share
[...] Read more.
Caspase (CASP) is a protease family that plays a vital role in apoptosis, development, and immune response. Herein, we reported the identification and characterization of two CASPs, AjCASPX1 and AjCASPX2, from the sea cucumber Apostichopus japonicus, an important aquaculture species. AjCASPX1/2 share similar domain organizations with the vertebrate initiator caspases CASP2/9, including the CARD domain and the p20/p10 subunits with conserved functional motifs. However, compared with human CASP2/9, AjCASPX1/2 possess unique structural features in the linker region between p20 and p10. AjCASPX1, but not AjCASPX2, induced marked apoptosis of human cells by activating CASP3/7. The recombinant proteins of AjCASPX2 and the CARD domain of AjCASPX2 were able to bind to a wide range of bacteria, as well as bacterial cell wall components, and inhibit bacterial growth. AjCASPX1, when expressed in Escherichia coli, was able to kill the host bacteria. Under normal conditions, AjCASPX1 and AjCASPX2 expressions were most abundant in sea cucumber muscle and coelomocytes, respectively. After bacterial infection, both AjCASPX1 and AjCASPX2 expressions were significantly upregulated in sea cucumber tissues and cells. Together, these results indicated that AjCASPX1 and AjCASPX2 were initiator caspases with antimicrobial activity and likely functioned in apoptosis and immune defense against pathogen infection.
Full article
(This article belongs to the Special Issue Genetics and Genomics in Aquatic Animals)
►▼
Show Figures
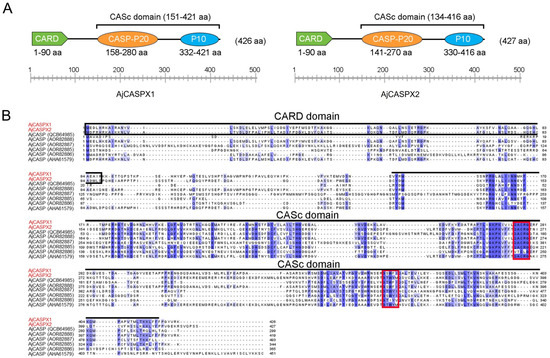
Figure 1
Open AccessReview
Oxidative Stress Biomarkers in Male Infertility: Established Methodologies and Future Perspectives
by
Filomena Mottola, Ilaria Palmieri, Maria Carannante, Angela Barretta, Shubhadeep Roychoudhury and Lucia Rocco
Genes 2024, 15(5), 539; https://doi.org/10.3390/genes15050539 - 25 Apr 2024
Abstract
Male fertility can be affected by oxidative stress (OS), which occurs when an imbalance between the production of reactive oxygen species (ROS) and the body’s ability to neutralize them arises. OS can damage cells and influence sperm production. High levels of lipid peroxidation
[...] Read more.
Male fertility can be affected by oxidative stress (OS), which occurs when an imbalance between the production of reactive oxygen species (ROS) and the body’s ability to neutralize them arises. OS can damage cells and influence sperm production. High levels of lipid peroxidation have been linked to reduced sperm motility and decreased fertilization ability. This literature review discusses the most commonly used biomarkers to measure sperm damage caused by ROS, such as the high level of OS in seminal plasma as an indicator of imbalance in antioxidant activity. The investigated biomarkers include 8-hydroxy-2-deoxyguanosine acid (8-OHdG), a marker of DNA damage caused by ROS, and F2 isoprostanoids (8-isoprostanes) produced by lipid peroxidation. Furthermore, this review focuses on recent methodologies including the NGS polymorphisms and differentially expressed gene (DEG) analysis, as well as the epigenetic mechanisms linked to ROS during spermatogenesis along with new methodologies developed to evaluate OS biomarkers. Finally, this review addresses a valuable insight into the mechanisms of male infertility provided by these advances and how they have led to new treatment possibilities. Overall, the use of biomarkers to evaluate OS in male infertility has supplied innovative diagnostic and therapeutic approaches, enhancing our understanding of male infertility mechanisms.
Full article
(This article belongs to the Section Cytogenomics)
►▼
Show Figures
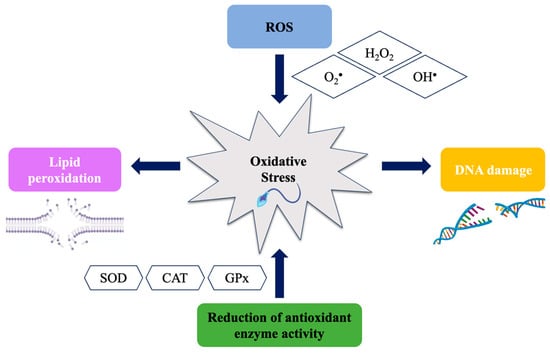
Figure 1
Open AccessReview
Colorectal Cancer: Genetic Underpinning and Molecular Therapeutics for Precision Medicine
by
Gideon T. Dosunmu and Ardaman Shergill
Genes 2024, 15(5), 538; https://doi.org/10.3390/genes15050538 - 25 Apr 2024
Abstract
Colorectal cancer (CRC) accounts for about 10% of all cancer cases and 9% of cancer-related deaths globally. In the United States alone, CRC represents approximately 12.6% of all cancer cases, with a mortality rate of about 8%. CRC is now the first leading
[...] Read more.
Colorectal cancer (CRC) accounts for about 10% of all cancer cases and 9% of cancer-related deaths globally. In the United States alone, CRC represents approximately 12.6% of all cancer cases, with a mortality rate of about 8%. CRC is now the first leading cause of cancer death in men younger than age 50 and second in women younger than age 50. This review delves into the genetic landscape of CRC, highlighting key mutations and their implications in disease progression and treatment. We provide an overview of the current and emerging therapeutic strategies tailored to individual genomic profiles.
Full article
(This article belongs to the Section Human Genomics and Genetic Diseases)
►▼
Show Figures
Figure 1

Journal Menu
► ▼ Journal Menu-
- Genes Home
- Aims & Scope
- Editorial Board
- Reviewer Board
- Topical Advisory Panel
- Instructions for Authors
- Special Issues
- Topics
- Sections & Collections
- Article Processing Charge
- Indexing & Archiving
- Editor’s Choice Articles
- Most Cited & Viewed
- Journal Statistics
- Journal History
- Journal Awards
- Society Collaborations
- Conferences
- Editorial Office
Journal Browser
► ▼ Journal BrowserHighly Accessed Articles
Latest Books
E-Mail Alert
News
Topics
Topic in
Biology, BioMed, Cells, Genes
Applications of the Zebrafish Model
Topic Editors: De-Li Shi, Pengfei XuDeadline: 30 June 2024
Topic in
Biomedicines, Cells, CIMB, Genes, IJMS
Animal Models of Human Disease 2.0
Topic Editors: Sigrun Lange, Jameel M. InalDeadline: 31 August 2024
Topic in
Agriculture, Bioengineering, Genes, IJMS, Plants
Genetic Engineering in Agriculture
Topic Editors: Amy L. Klocko, Jianjun Chen, Haiwei LuDeadline: 30 September 2024
Topic in
Biology, BioMedInformatics, Cancers, Genes, IJMS
The 22nd International Conference on Bioinformatics (InCoB 2023): Translational Bioinformatics Transforming Life
Topic Editors: Jyotsna Batra, Srilakshmi Srinivasan, Shoba Ranganathan, Asif M. Khan, Harpreet SinghDeadline: 15 November 2024

Conferences
Special Issues
Special Issue in
Genes
Cotton Genes, Genetics, and Genomics
Guest Editor: Guanjing HuDeadline: 10 May 2024
Special Issue in
Genes
DNA Damage and Repair in Microorganisms, Plants and Mammalian Systems
Guest Editors: Ioly Kotta-Loizou, Nan Zhang, Xin WangDeadline: 20 May 2024
Special Issue in
Genes
Commemorating the Launch of the Section "Cytogenomics"
Guest Editor: Darren GriffinDeadline: 5 June 2024
Special Issue in
Genes
Genetic Markers and Liquid Biopsy for Kidney Diseases
Guest Editors: Guorong Li, Christophe MariatDeadline: 15 June 2024
Topical Collections
Topical Collection in
Genes
Study on Genotypes and Phenotypes of Pediatric Clinical Rare Diseases
Collection Editors: Livia Garavelli, Stefano Giuseppe Caraffi
Topical Collection in
Genes
Eukaryotic Non-coding RNAs: Diversity, Structure/Function, Implication in Cardiovascular Disease
Collection Editors: Morten Andre Høydal, Christiane Branlant
Topical Collection in
Genes
Feature Papers in ‘Animal Genetics and Genomics’
Collection Editors: Antonio Figueras, Raquel Vasconcelos









