Journal Description
Viruses
Viruses
is a peer-reviewed, open access journal of virology, published monthly online by MDPI. The American Society for Virology (ASV), Spanish Society for Virology (SEV), Canadian Society for Virology (CSV), Italian Society for Virology (SIV-ISV), Australasian Virology Society (AVS) and others are affiliated with Viruses and their members receive a discount on the article processing charges.
- Open Access— free for readers, with article processing charges (APC) paid by authors or their institutions.
- High Visibility: indexed within Scopus, SCIE (Web of Science), PubMed, MEDLINE, PMC, Embase, PubAg, AGRIS, and other databases.
- Journal Rank: JCR - Q2 (Virology) / CiteScore - Q1 (Infectious Diseases)
- Rapid Publication: manuscripts are peer-reviewed and a first decision is provided to authors approximately 13.8 days after submission; acceptance to publication is undertaken in 2.5 days (median values for papers published in this journal in the second half of 2023).
- Recognition of Reviewers: reviewers who provide timely, thorough peer-review reports receive vouchers entitling them to a discount on the APC of their next publication in any MDPI journal, in appreciation of the work done.
- Companion journals for Viruses include: COVID and Zoonotic Diseases.
Impact Factor:
4.7 (2022);
5-Year Impact Factor:
4.8 (2022)
Latest Articles
Diagnostic Utility of Cytomegalovirus (CMV) DNA Quantitation in Ulcerative Colitis
Viruses 2024, 16(5), 691; https://doi.org/10.3390/v16050691 (registering DOI) - 26 Apr 2024
Abstract
Cytomegalovirus (CMV) colitis is a critical condition associated with severe complications in ulcerative colitis (UC). This study aimed to investigate the diagnostic value of the presence of CMV DNA in intestinal mucosa tissue and blood samples in patients with active UC. This study
[...] Read more.
Cytomegalovirus (CMV) colitis is a critical condition associated with severe complications in ulcerative colitis (UC). This study aimed to investigate the diagnostic value of the presence of CMV DNA in intestinal mucosa tissue and blood samples in patients with active UC. This study included 81 patients with exacerbated symptoms of UC. Patient data were obtained from the Hospital Information Management System. CMV DNA in colorectal tissue and plasma samples were analyzed using a real-time quantitative PCR assay. CMV markers were detected using immunohistochemistry and hematoxylin–eosin staining. Immunohistochemistry positivity was observed in tissue samples from eight (9.9%) patients. Only one (1.2%) patient showed CMV-specific intranuclear inclusion bodies. CMV DNA was detected in 63.0% of the tissues (median: 113 copies/mg) and in 58.5% of the plasma samples (median: 102 copies/mL). For tissues, sensitivity and the negative predictive value (NPV) for qPCR were excellent (100.0%), whereas specificity and the positive predictive value (PPV) were low (41.9% and 15.7%, respectively). For plasma, sensitivity and NPV were high (100.0%) for qPCR, whereas specificity and PPV were low (48.6% and 24.0%, respectively). CMV DNA ≥392 copies/mg in tissue samples (sensitivity 100.0% and specificity 83.6%) and ≥578 copies/mL (895 IU/mL) in plasma samples (sensitivity 66.7% and specificity 100.0%) provided an optimal diagnosis for this test. The qPCR method improved patient management through the early detection of CMV colitis in patients with UC. However, reliance on qPCR positivity alone can lead to overdiagnosis. Quantification of CMV DNA can improve diagnostic specificity, although standardization is warranted.
Full article
(This article belongs to the Section Human Virology and Viral Diseases)
Open AccessReview
Challenges in Treating Pediatric Cancer Patients during the COVID-19 Pandemic: Balancing Risks and Care
by
Juan Luis Chávez-Pacheco, Manuel Castillejos-López, Laura M. Hernández-Regino, Liliana Velasco-Hidalgo, Marta Zapata-Tarres, Valeria Correa-Carranza, Guillermo Rosario-Méndez, Rehotbevely Barrientos-Ríos, Arnoldo Aquino-Gálvez and Luz María Torres-Espíndola
Viruses 2024, 16(5), 690; https://doi.org/10.3390/v16050690 - 26 Apr 2024
Abstract
The COVID-19 pandemic has resulted in millions of fatalities worldwide. The case of pediatric cancer patients stands out since, despite being considered a population at risk, few studies have been carried out concerning symptom detection or the description of the mechanisms capable of
[...] Read more.
The COVID-19 pandemic has resulted in millions of fatalities worldwide. The case of pediatric cancer patients stands out since, despite being considered a population at risk, few studies have been carried out concerning symptom detection or the description of the mechanisms capable of modifying the course of the COVID-19 disease, such as the interaction and response between the virus and the treatment given to cancer patients. By synthesizing existing studies, this paper aims to expose the treatment challenges for pediatric patients with COVID-19 in an oncology context. Additionally, this updated review includes studies that utilized the antiviral agents Remdesivir and PaxlovidTM in pediatric cancer patients. There is no specific treatment designed exclusively for pediatric cancer patients dealing with COVID-19, and it is advisable to avoid self-medication to prevent potential side effects. Managing COVID-19 in pediatric cancer patients is indeed a substantial challenge. New strategies, such as chemotherapy application rooms, have been implemented for children with cancer who were positive for COVID-19 but asymptomatic since the risk of disease progression is greater than the risk of complications from SARS-CoV-2.
Full article
(This article belongs to the Section Coronaviruses)
Open AccessArticle
Deletion of 82–85 N-Terminal Residues in SARS-CoV-2 Nsp1 Restricts Virus Replication
by
Gianni Gori Savellini, Gabriele Anichini, Fabrizio Manetti, Claudia Immacolata Trivisani and Maria Grazia Cusi
Viruses 2024, 16(5), 689; https://doi.org/10.3390/v16050689 - 26 Apr 2024
Abstract
Non-structural protein 1 (Nsp1) represents one of the most crucial SARS-CoV-2 virulence factors by inhibiting the translation of host mRNAs and promoting their degradation. We selected naturally occurring virus lineages with specific Nsp1 deletions located at both the N- and C-terminus of the
[...] Read more.
Non-structural protein 1 (Nsp1) represents one of the most crucial SARS-CoV-2 virulence factors by inhibiting the translation of host mRNAs and promoting their degradation. We selected naturally occurring virus lineages with specific Nsp1 deletions located at both the N- and C-terminus of the protein. Our data provide new insights into how Nsp1 coordinates these functions on host and viral mRNA recognition. Residues 82–85 in the N-terminal part of Nsp1 likely play a role in docking the 40S mRNA entry channel, preserving the inhibition of host gene expression without affecting cellular mRNA decay. Furthermore, this domain prevents viral mRNAs containing the 5′-leader sequence to escape translational repression. These findings support the presence of distinct domains within the Nsp1 protein that differentially modulate mRNA recognition, translation and turnover. These insights have implications for the development of drugs targeting viral proteins and provides new evidences of how specific mutations in SARS-CoV-2 Nsp1 could attenuate the virus.
Full article
(This article belongs to the Section Coronaviruses)
►▼
Show Figures
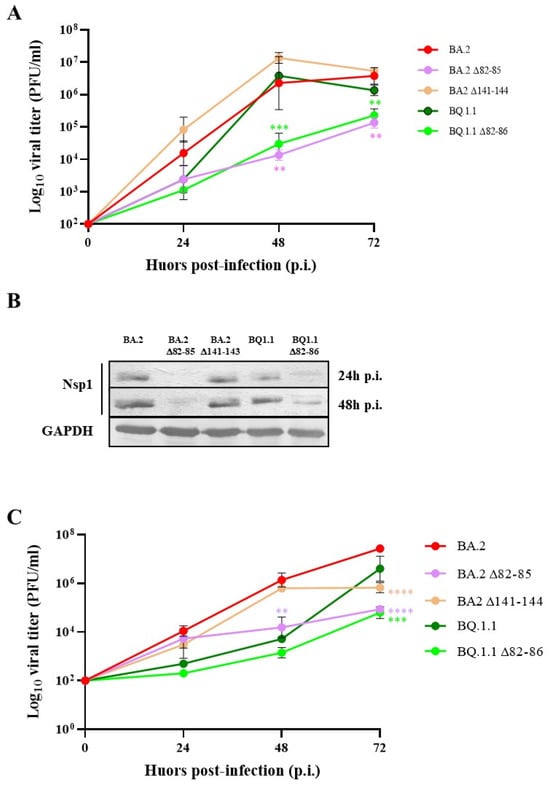
Figure 1
Open AccessArticle
Long-Term Impairment of Working Ability in Subjects under 60 Years of Age Hospitalised for COVID-19 at 2 Years of Follow-Up: A Cross-Sectional Study
by
Luisa Frallonardo, Annunziata Ilenia Ritacco, Angela Amendolara, Domenica Cassano, Giorgia Manco Cesari, Alessia Lugli, Mariangela Cormio, Michele De Filippis, Greta Romita, Giacomo Guido, Luigi Piccolomo, Vincenzo Giliberti, Francesco Cavallin, Francesco Vladimiro Segala, Francesco Di Gennaro and Annalisa Saracino
Viruses 2024, 16(5), 688; https://doi.org/10.3390/v16050688 - 26 Apr 2024
Abstract
Coronavirus disease 2019 (COVID-19) can lead to persistent and debilitating symptoms referred to as Post-Acute sequelae of SARS-CoV-2 infection (PASC) This broad symptomatology lasts for months after the acute infection and impacts physical and mental health and everyday functioning. In the present study,
[...] Read more.
Coronavirus disease 2019 (COVID-19) can lead to persistent and debilitating symptoms referred to as Post-Acute sequelae of SARS-CoV-2 infection (PASC) This broad symptomatology lasts for months after the acute infection and impacts physical and mental health and everyday functioning. In the present study, we aimed to evaluate the prevalence and predictors of long-term impairment of working ability in non-elderly people hospitalised for COVID-19. Methods: This cross-sectional study involved 322 subjects hospitalised for COVID-19 from 1 March 2020 to 31 December 2022 in the University Hospital of Bari, Apulia, Italy, enrolled at the time of their hospital discharge and followed-up at a median of 731 days since hospitalization (IQR 466–884). Subjects reporting comparable working ability and those reporting impaired working ability were compared using the Mann-Whitney test (continuous data) and Fisher’s test or Chi-Square test (categorical data). Multivariable analysis of impaired working ability was performed using a logistic regression model. Results: Among the 322 subjects who were interviewed, 184 reported comparable working ability (57.1%) and 134 reported impaired working ability (41.6%) compared to the pre-COVID-19 period. Multivariable analysis identified age at hospital admission (OR 1.02, 95% CI 0.99 to 1.04), female sex (OR 1.90, 95% CI 1.18 to 3.08), diabetes (OR 3.73, 95% CI 1.57 to 9.65), receiving oxygen during hospital stay (OR 1.76, 95% CI 1.01 to 3.06), and severe disease (OR 0.51, 95% CI 0.26 to 1.01) as independent predictors of long-term impaired working ability after being hospitalised for COVID-19. Conclusions: Our findings suggest that PASC promotes conditions that could result in decreased working ability and unemployment. These results highlight the significant impact of this syndrome on public health and the global economy, and the need to develop clinical pathways and guidelines for long-term care with specific focus on working impairment.
Full article
(This article belongs to the Section Human Virology and Viral Diseases)
Open AccessArticle
Development of Attenuated Viruses for Effective Protection against Pepper Veinal Mottle Virus in Tomato Crops
by
Guan-Da Wang, Chian-Chi Lin and Tsung-Chi Chen
Viruses 2024, 16(5), 687; https://doi.org/10.3390/v16050687 - 26 Apr 2024
Abstract
Tomato (Solanum lycopersicum) is the most important vegetable and fruit crop in the family Solanaceae worldwide. Numerous pests and pathogens, especially viruses, severely affect tomato production, causing immeasurable market losses. In Taiwan, the cultivation of tomato crops is mainly threatened by
[...] Read more.
Tomato (Solanum lycopersicum) is the most important vegetable and fruit crop in the family Solanaceae worldwide. Numerous pests and pathogens, especially viruses, severely affect tomato production, causing immeasurable market losses. In Taiwan, the cultivation of tomato crops is mainly threatened by insect-borne viruses, among which pepper veinal mottle virus (PVMV) is one of the most prevalent. PVMV is a member of the genus Potyvirus of the family Potyviridae and is non-persistently transmitted by aphids. Its infection significantly reduces tomato fruit yield and quality. So far, no PVMV-resistant tomato lines are available. In this study, we performed nitrite-induced mutagenesis of the PVMV tomato isolate Tn to generate attenuated PVMV mutants. PVMV Tn causes necrotic lesions in Chenopodium quinoa leaves and severe mosaic and wilting in Nicotiana benthamiana plants. After nitrite treatment, three attenuated PVMV mutants, m4-8, m10-1, and m10-11, were selected while inducing milder responses to C. quinoa and N. benthamiana with lower accumulation in tomato plants. In greenhouse tests, the three mutants showed different degrees of cross-protection against wild-type PVMV Tn. m4-8 showed the highest protective efficacy against PVMV Tn in N. benthamiana and tomato plants, 100% and 97.9%, respectively. A whole-genome sequence comparison of PVMV Tn and m4-8 revealed that 20 nucleotide substitutions occurred in the m4-8 genome, resulting in 18 amino acid changes. Our results suggest that m4-8 has excellent potential to protect tomato crops from PVMV. The application of m4-8 in protecting other Solanaceae crops, such as peppers, will be studied in the future.
Full article
(This article belongs to the Special Issue Crop Resistance to Viral Infections)
Open AccessArticle
Unraveling the Properties of Phage Display Fab Libraries and Their Use in the Selection of Gliadin-Specific Probes by Applying High-Throughput Nanopore Sequencing
by
Eduardo Garcia-Calvo, Aina García-García, Santiago Rodríguez, Rosario Martín and Teresa García
Viruses 2024, 16(5), 686; https://doi.org/10.3390/v16050686 - 26 Apr 2024
Abstract
Directed evolution is a pivotal strategy for new antibody discovery, which allowed the generation of high-affinity Fabs against gliadin from two antibody libraries in our previous studies. One of the libraries was exclusively derived from celiac patients’ mRNA (immune library) while the other
[...] Read more.
Directed evolution is a pivotal strategy for new antibody discovery, which allowed the generation of high-affinity Fabs against gliadin from two antibody libraries in our previous studies. One of the libraries was exclusively derived from celiac patients’ mRNA (immune library) while the other was obtained through a protein engineering approach (semi-immune library). Recent advances in high-throughput DNA sequencing techniques are revolutionizing research across genomics, epigenomics, and transcriptomics. In the present work, an Oxford Nanopore in-lab sequencing device was used to comprehensively characterize the composition of the constructed libraries, both at the beginning and throughout the phage-mediated selection processes against gliadin. A customized analysis pipeline was used to select high-quality reads, annotate chain distribution, perform sequence analysis, and conduct statistical comparisons between the different selection rounds. Some immunological attributes of the most representative phage variants after the selection process were also determined. Sequencing results revealed the successful transfer of the celiac immune response features to the immune library and the antibodies derived from it, suggesting the crucial role of these features in guiding the selection of high-affinity recombinant Fabs against gliadin. In summary, high-throughput DNA sequencing has improved our understanding of the selection processes aimed at generating molecular binders against gliadin.
Full article
(This article belongs to the Special Issue Biotechnological Applications of Phage and Phage-Derived Proteins 4.0)
►▼
Show Figures
Figure 1
Open AccessReview
Impact of Prior COVID-19 Immunization and/or Prior Infection on Immune Responses and Clinical Outcomes
by
Achilleas Livieratos, Charalambos Gogos and Karolina Akinosoglou
Viruses 2024, 16(5), 685; https://doi.org/10.3390/v16050685 - 26 Apr 2024
Abstract
Cellular and humoral immunity exhibit dynamic adaptation to the mutating SARS-CoV-2 virus. It is noteworthy that immune responses differ significantly, influenced by whether a patient has received vaccination or whether there is co-occurrence of naturally acquired and vaccine-induced immunity, known as hybrid immunity.
[...] Read more.
Cellular and humoral immunity exhibit dynamic adaptation to the mutating SARS-CoV-2 virus. It is noteworthy that immune responses differ significantly, influenced by whether a patient has received vaccination or whether there is co-occurrence of naturally acquired and vaccine-induced immunity, known as hybrid immunity. The different immune reactions, conditional on vaccination status and the viral variant involved, bear implications for inflammatory responses, patient outcomes, pathogen transmission rates, and lingering post-COVID conditions. Considering these developments, we have performed a review of recently published literature, aiming to disentangle the intricate relationships among immunological profiles, transmission, the long-term health effects post-COVID infection poses, and the resultant clinical manifestations. This investigation is directed toward understanding the variability in the longevity and potency of cellular and humoral immune responses elicited by immunization and hybrid infection.
Full article
(This article belongs to the Special Issue Viral Sepsis: Pathogenesis, Diagnostics and Therapeutics)
►▼
Show Figures
Figure 1
Open AccessReview
Hepatitis E Virus in Domestic Ruminants and Virus Excretion in Milk—A Potential Source of Zoonotic HEV Infection
by
Gergana Zahmanova, Katerina Takova, Georgi L. Lukov and Anton Andonov
Viruses 2024, 16(5), 684; https://doi.org/10.3390/v16050684 - 26 Apr 2024
Abstract
The hepatitis E virus is a serious health concern worldwide, with 20 million cases each year. Growing numbers of autochthonous HEV infections in industrialized nations are brought on via the zoonotic transmission of HEV genotypes 3 and 4. Pigs and wild boars are
[...] Read more.
The hepatitis E virus is a serious health concern worldwide, with 20 million cases each year. Growing numbers of autochthonous HEV infections in industrialized nations are brought on via the zoonotic transmission of HEV genotypes 3 and 4. Pigs and wild boars are the main animal reservoirs of HEV and play the primary role in HEV transmission. Consumption of raw or undercooked pork meat and close contact with infected animals are the most common causes of hepatitis E infection in industrialized countries. However, during the past few years, mounting data describing HEV distribution has led experts to believe that additional animals, particularly domestic ruminant species (cow, goat, sheep, deer, buffalo, and yak), may also play a role in the spreading of HEV. Up to now, there have not been enough studies focused on HEV infections associated with animal milk and the impact that they could have on the epidemiology of HEV. This critical analysis discusses the role of domestic ruminants in zoonotic HEV transmissions. More specifically, we focus on concerns related to milk safety, the role of mixed farming in cross-species HEV infections, and what potential consequences these may have on public health.
Full article
(This article belongs to the Special Issue Hepatitis E: Molecular Virology, Pathogenesis, and Treatment)
►▼
Show Figures

Figure 1
Open AccessArticle
Evolution Characterization and Pathogenicity of an NADC34-like PRRSV Isolated from Inner Mongolia, China
by
Hong-Zhe Zhao, Chun-Yu Liu, Hai Meng, Cheng-Long Sun, Hong-Wen Yang, Hao Wang, Jian Zou, Peng Li, Feng-Ye Han, Gen Qi, Yang Zhang, Bing-Bing Lin, Chuang Liu, Meng-Meng Chen, Pan-Ling Zhang, Xiao-Dong Chen, Yi-Di Zhang, Qian-Jin Song, Yong-Jun Wen and Feng-Xue Wang
Viruses 2024, 16(5), 683; https://doi.org/10.3390/v16050683 - 26 Apr 2024
Abstract
Porcine reproductive and respiratory syndrome virus (PRRSV) is a pathogen that causes severe abortions in sows and high piglet mortality, resulting in huge economic losses to the pig industry worldwide. The emerging and novel PRRSV isolates are clinically and biologically important, as there
[...] Read more.
Porcine reproductive and respiratory syndrome virus (PRRSV) is a pathogen that causes severe abortions in sows and high piglet mortality, resulting in huge economic losses to the pig industry worldwide. The emerging and novel PRRSV isolates are clinically and biologically important, as there are likely recombination and pathogenic differences among PRRSV genomes. Furthermore, the NADC34-like strain has become a major epidemic strain in some parts of China, but the characterization and pathogenicity of the latest strain in Inner Mongolia have not been reported in detail. In this study, an NADC34-like strain (CHNMGKL1-2304) from Tongliao City, Inner Mongolia was successfully isolated and characterized, and confirmed the pathogenicity in pigs. The phylogenetic tree showed that this strain belonged to sublineage 1.5 and had high homology with the strain JS2021NADC34. There is no recombination between CHNMGKL1-2304 and any other domestic strains. Animal experiments show that the CHNMGKL1-2304 strain is moderately virulent to piglets, which show persistent fever, weight loss and high morbidity but no mortality. The presence of PRRSV nucleic acids was detected in both blood, tissues, nasal and fecal swabs. In addition, obvious pathological changes and positive signals were observed in lung, lymph node, liver and spleen tissues when subjected to hematoxylin–eosin (HE) staining and immunohistochemistry (IHC). This report can provide a basis for epidemiological investigations and subsequent studies of PRRSV.
Full article
(This article belongs to the Section Animal Viruses)
►▼
Show Figures
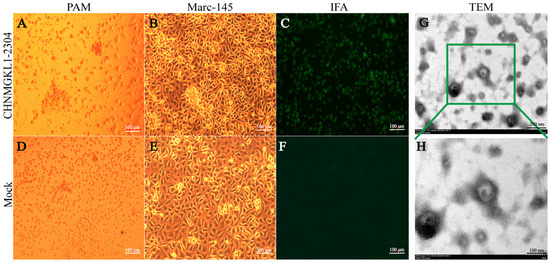
Figure 1
Open AccessArticle
Reduction of the Risk of Hepatocellular Carcinoma over Time Using Direct-Acting Antivirals: A Propensity Score Analysis of a Real-Life Cohort (PITER HCV)
by
Maria Giovanna Quaranta, Luisa Cavalletto, Francesco Paolo Russo, Vincenza Calvaruso, Luigina Ferrigno, Alberto Zanetto, Benedetta Mattioli, Roberta D’Ambrosio, Valentina Panetta, Giuseppina Brancaccio, Giovanni Raimondo, Maurizia Rossana Brunetto, Anna Linda Zignego, Carmine Coppola, Andrea Iannone, Elisa Biliotti, Elena Rosselli Del Turco, Marco Massari, Anna Licata, Francesco Barbaro, Marcello Persico, Filomena Morisco, Maurizio Pompili, Federica Cerini, Massimo Puoti, Teresa Santantonio, Antonio Craxì, Loreta A. Kondili, Liliana Chemello Investigators and on behalf of PITER Collaborating Investigatorsadd
Show full author list
remove
Hide full author list
Viruses 2024, 16(5), 682; https://doi.org/10.3390/v16050682 - 26 Apr 2024
Abstract
The treatment of hepatitis C virus (HCV) with direct-acting antivirals (DAA) leads to high sustained virological response (SVR) rates, but hepatocellular carcinoma (HCC) risk persists in people with advanced liver disease even after SVR. We weighted the HCC risk in people with cirrhosis
[...] Read more.
The treatment of hepatitis C virus (HCV) with direct-acting antivirals (DAA) leads to high sustained virological response (SVR) rates, but hepatocellular carcinoma (HCC) risk persists in people with advanced liver disease even after SVR. We weighted the HCC risk in people with cirrhosis achieving HCV eradication through DAA treatment and compared it with untreated participants in the multicenter prospective Italian Platform for the Study of Viral Hepatitis Therapies (PITER) cohort. Propensity matching with inverse probability weighting was used to compare DAA-treated and untreated HCV-infected participants with liver cirrhosis. Kaplan–Meier analysis and competing risk regression analysis were performed. Within the first 36 months, 30 de novo HCC cases occurred in the untreated group (n = 307), with a weighted incidence rate of 0.34% (95%CI: 0.23–0.52%), compared to 63 cases among SVR patients (n = 1111), with an incidence rate of 0.20% (95%CI: 0.16–0.26%). The 12-, 24-, and 36-month HCC weighted cumulative incidence rates were 6.7%, 8.4%, and 10.0% in untreated cases and 2.3%, 4.5%, and 7.0% in the SVR group. Considering death or liver transplantation as competing events, the untreated group showed a 64% higher risk of HCC incidence compared to SVR patients (SubHR 1.64, 95%CI: 1.02–2.62). Other variables independently associated with the HCC occurrence were male sex, increasing age, current alcohol use, HCV genotype 3, platelet count ≤ 120,000/µL, and albumin ≤ 3.5 g/dL. In real-life practice, the high efficacy of DAA in achieving SVR is translated into high effectiveness in reducing the HCC incidence risk.
Full article
(This article belongs to the Special Issue Recent Advances in Anti-HCV, Anti-HBV and Anti-flavivirus Agents)
►▼
Show Figures
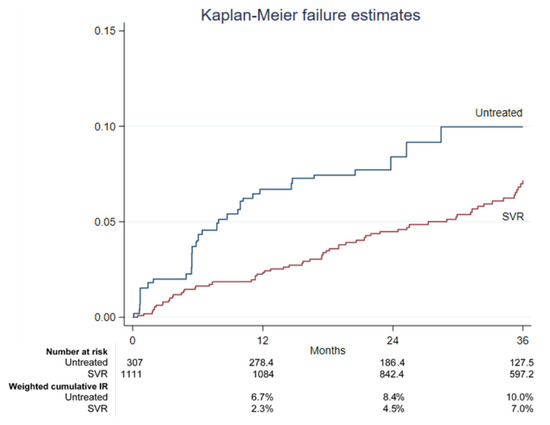
Figure 1
Open AccessArticle
Monkeypox Virus Neutralizing Antibodies at Six Months from Mpox Infection: Virologic Factors Associated with Poor Immunologic Response
by
Angelo Roberto Raccagni, Alessandro Mancon, Sara Diotallevi, Riccardo Lolatto, Elena Bruzzesi, Maria Rita Gismondo, Antonella Castagna, Davide Mileto and Silvia Nozza
Viruses 2024, 16(5), 681; https://doi.org/10.3390/v16050681 - 26 Apr 2024
Abstract
A natural monkeypox virus infection may not induce sufficient neutralizing antibody responses in a subset of healthy individuals. The aim of this study was to evaluate monkeypox virus-neutralizing antibodies six months after infection and to assess the virological factors predictive of a poor
[...] Read more.
A natural monkeypox virus infection may not induce sufficient neutralizing antibody responses in a subset of healthy individuals. The aim of this study was to evaluate monkeypox virus-neutralizing antibodies six months after infection and to assess the virological factors predictive of a poor immunological response. Antibodies were assessed using a plaque reduction neutralization test at six months from mpox infection; mpox cutaneous, oropharyngeal, and anal swabs, semen, and plasma samples were tested during infection. Overall, 95 people were included in the study; all developed detectable antibodies. People who were positive for the monkeypox virus for more days had higher levels of antibodies when considering all tested samples (p = 0.029) and all swabs (p = 0.005). Mpox cycle threshold values were not predictive of antibody titers. This study found that the overall days of monkeypox virus detection in the body, irrespective of the viral loads, were directly correlated with monkeypox virus neutralizing antibodies at six months after infection.
Full article
(This article belongs to the Section Viral Immunology, Vaccines, and Antivirals)
►▼
Show Figures
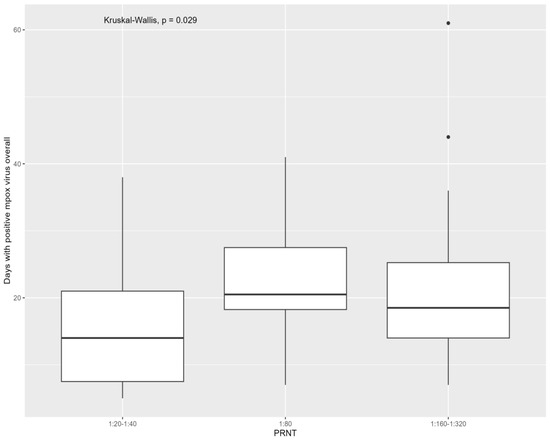
Figure 1
Open AccessReview
Human Papillomavirus and Associated Cancers: A Review
by
JaNiese E. Jensen, Greta L. Becker, J. Brooks Jackson and Mary B. Rysavy
Viruses 2024, 16(5), 680; https://doi.org/10.3390/v16050680 - 26 Apr 2024
Abstract
The human papillomavirus is the most common sexually transmitted infection in the world. Most HPV infections clear spontaneously within 2 years of infection; however, persistent infection can result in a wide array of diseases, ranging from genital warts to cancer. Most cases of
[...] Read more.
The human papillomavirus is the most common sexually transmitted infection in the world. Most HPV infections clear spontaneously within 2 years of infection; however, persistent infection can result in a wide array of diseases, ranging from genital warts to cancer. Most cases of cervical, anal, and oropharyngeal cancers are due to HPV infection, with cervical cancer being one of the leading causes of cancer death in women worldwide. Screening is available for HPV and cervical cancer, but is not available everywhere, particularly in lower-resource settings. HPV infection disproportionally affects individuals living with HIV, resulting in decreased clearance, increased development of cancer, and increased mortality. The development of the HPV vaccine has shown a drastic decrease in HPV-related diseases. The vaccine prevents cervical cancer with near 100% efficacy, if given prior to first sexual activity. Vaccination uptake remains low worldwide due to a lack of access and limited knowledge of HPV. Increasing awareness of HPV and access to vaccination are necessary to decrease cancer and HPV-related morbidity and mortality worldwide.
Full article
(This article belongs to the Special Issue Chronic Infection by Oncogenic Viruses)
►▼
Show Figures
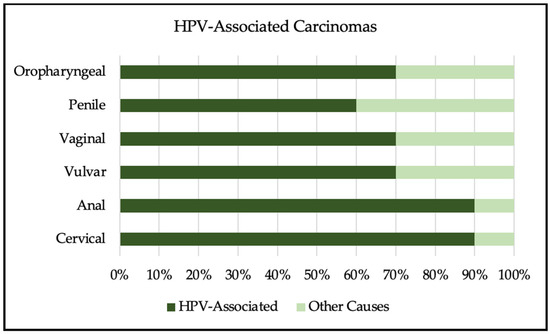
Figure 1
Open AccessReview
T Cell Surveillance during Cutaneous Viral Infections
by
Luxin Pei and Heather D. Hickman
Viruses 2024, 16(5), 679; https://doi.org/10.3390/v16050679 - 26 Apr 2024
Abstract
The skin is a complex tissue that provides a strong physical barrier against invading pathogens. Despite this, many viruses can access the skin and successfully replicate in either the epidermal keratinocytes or dermal immune cells. In this review, we provide an overview of
[...] Read more.
The skin is a complex tissue that provides a strong physical barrier against invading pathogens. Despite this, many viruses can access the skin and successfully replicate in either the epidermal keratinocytes or dermal immune cells. In this review, we provide an overview of the antiviral T cell biology responding to cutaneous viral infections and how these responses differ depending on the cellular targets of infection. Much of our mechanistic understanding of T cell surveillance of cutaneous infection has been gained from murine models of poxvirus and herpesvirus infection. However, we also discuss other viral infections, including flaviviruses and papillomaviruses, in which the cutaneous T cell response has been less extensively studied. In addition to the mechanisms of successful T cell control of cutaneous viral infection, we highlight knowledge gaps and future directions with possible impact on human health.
Full article
(This article belongs to the Special Issue Innate and Adaptive Immunity to Cutaneous Virus Infection)
►▼
Show Figures
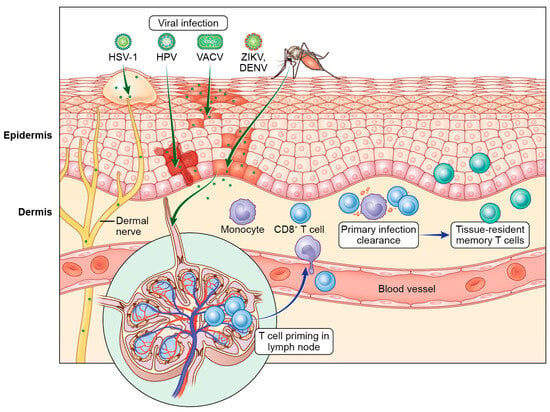
Figure 1
Open AccessArticle
Innate Immune Evasion of PRRSV nsp11 through Degradation of the HDAC2 by Its Endoribonuclease Activity
by
He Zhang, Jianxing Chen, Changqing Yu, Yu Pan, Wenjie Ma, Hao Feng, Jinxin Xie, Hongyan Chen, Yue Wang and Changyou Xia
Viruses 2024, 16(5), 678; https://doi.org/10.3390/v16050678 - 25 Apr 2024
Abstract
Porcine reproductive and respiratory syndrome virus (PRRSV), a member of the Arteriviridae family, represents a persistent menace to the global pig industry, causing reproductive failure and respiratory disease in pigs. In this study, we delved into the role of histone deacetylases (HDAC2) during
[...] Read more.
Porcine reproductive and respiratory syndrome virus (PRRSV), a member of the Arteriviridae family, represents a persistent menace to the global pig industry, causing reproductive failure and respiratory disease in pigs. In this study, we delved into the role of histone deacetylases (HDAC2) during PRRSV infection. Our findings revealed that HDAC2 expression is downregulated upon PRRSV infection. Notably, suppressing HDAC2 activity through specific small interfering RNA led to an increase in virus production, whereas overexpressing HDAC2 effectively inhibited PRRSV replication by boosting the expression of IFN-regulated antiviral molecules. Furthermore, we identified the virus’s nonstructural protein 11 (nsp11) as a key player in reducing HDAC2 levels. Mutagenic analyses of PRRSV nsp11 revealed that its antagonistic effect on the antiviral activity of HDAC2 is dependent on its endonuclease activity. In summary, our research uncovered a novel immune evasion mechanism employed by PRRSV, providing crucial insights into the pathogenesis of this virus and guiding the development of innovative prevention strategies against PRRSV infection.
Full article
(This article belongs to the Section Viral Immunology, Vaccines, and Antivirals)
►▼
Show Figures
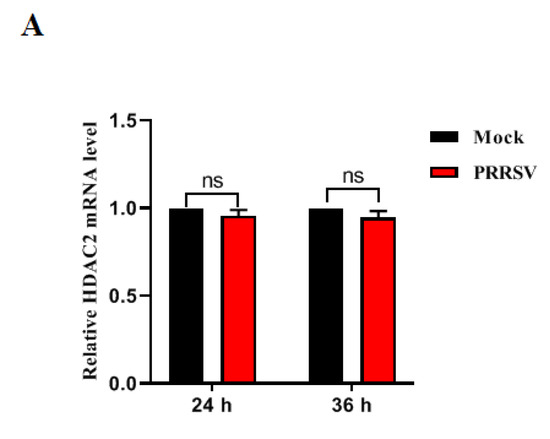
Figure 1
Open AccessArticle
Caboxamycin Inhibits Heart Inflammation in a Coxsackievirus B3-Induced Myocarditis Mouse Model
by
Hong-Gi Kim, Prima F. Hillman, You-Jeung Lee, Ha-Eun Jeon, Byung-Kwan Lim and Sang-Jip Nam
Viruses 2024, 16(5), 677; https://doi.org/10.3390/v16050677 - 25 Apr 2024
Abstract
Coxsackievirus B3 (CVB3) is a positive single-strand RNA genome virus which belongs to the enterovirus genus in the picornavirus family, like poliovirus. It is one of the most prevalent pathogens that cause myocarditis and pancreatitis in humans. However, a suitable therapeutic medication and
[...] Read more.
Coxsackievirus B3 (CVB3) is a positive single-strand RNA genome virus which belongs to the enterovirus genus in the picornavirus family, like poliovirus. It is one of the most prevalent pathogens that cause myocarditis and pancreatitis in humans. However, a suitable therapeutic medication and vaccination have yet to be discovered. Caboxamycin, a benzoxazole antibiotic isolated from the culture broth of the marine strain Streptomyces sp., SC0774, showed an antiviral effect in CVB3-infected HeLa cells and a CVB3-induced myocarditis mouse model. Caboxamycin substantially decreased CVB3 VP1 production and cleavage of translation factor eIF4G1 from CVB3 infection. Virus-positive and -negative strand RNA was dramatically reduced by caboxamycin treatment. In addition, the cleavage of the pro-apoptotic molecules BAD, BAX, and caspase3 was significantly inhibited by caboxamycin treatment. In animal experiments, the survival rate of mice was improved following caboxamycin treatment. Moreover, caboxamycin treatment significantly decreased myocardial damage and inflammatory cell infiltration. Our study showed that caboxamycin dramatically suppressed cardiac inflammation and mouse death. This result suggests that caboxamycin may be suitable as a potential antiviral drug for CVB3.
Full article
(This article belongs to the Special Issue An Update on Enterovirus Research)
Open AccessArticle
Investigating the Interactions of the Cucumber Mosaic Virus 2b Protein with the Viral 1a Replicase Component and the Cellular RNA Silencing Factor Argonaute 1
by
Sam Crawshaw, Alex M. Murphy, Pamela J. E. Rowling, Daniel Nietlispach, Laura S. Itzhaki and John P. Carr
Viruses 2024, 16(5), 676; https://doi.org/10.3390/v16050676 - 25 Apr 2024
Abstract
The cucumber mosaic virus (CMV) 2b protein is a suppressor of plant defenses and a pathogenicity determinant. Amongst the 2b protein’s host targets is the RNA silencing factor Argonaute 1 (AGO1), which it binds to and inhibits. In Arabidopsis thaliana, if 2b-induced
[...] Read more.
The cucumber mosaic virus (CMV) 2b protein is a suppressor of plant defenses and a pathogenicity determinant. Amongst the 2b protein’s host targets is the RNA silencing factor Argonaute 1 (AGO1), which it binds to and inhibits. In Arabidopsis thaliana, if 2b-induced inhibition of AGO1 is too efficient, it induces reinforcement of antiviral silencing by AGO2 and triggers increased resistance against aphids, CMV’s insect vectors. These effects would be deleterious to CMV replication and transmission, respectively, but are moderated by the CMV 1a protein, which sequesters sufficient 2b protein molecules into P-bodies to prevent excessive inhibition of AGO1. Mutant 2b protein variants were generated, and red and green fluorescent protein fusions were used to investigate subcellular colocalization with AGO1 and the 1a protein. The effects of mutations on complex formation with the 1a protein and AGO1 were investigated using bimolecular fluorescence complementation and co-immunoprecipitation assays. Although we found that residues 56–60 influenced the 2b protein’s interactions with the 1a protein and AGO1, it appears unlikely that any single residue or sequence domain is solely responsible. In silico predictions of intrinsic disorder within the 2b protein secondary structure were supported by circular dichroism (CD) but not by nuclear magnetic resonance (NMR) spectroscopy. Intrinsic disorder provides a plausible model to explain the 2b protein’s ability to interact with AGO1, the 1a protein, and other factors. However, the reasons for the conflicting conclusions provided by CD and NMR must first be resolved.
Full article
(This article belongs to the Special Issue Plant Viruses: Pirates of Cellular Pathways, 2nd Edition)
Open AccessArticle
First Report of Endemic Frog Virus 3 (FV3)-like Ranaviruses in the Korean Clawed Salamander (Onychodactylus koreanus) in Asia
by
Jongsun Kim, Haan Woo Sung, Tae Sung Jung, Jaejin Park and Daesik Park
Viruses 2024, 16(5), 675; https://doi.org/10.3390/v16050675 - 25 Apr 2024
Abstract
Frog virus 3 (FV3) in the genus Ranavirus of the family Iridoviridae causes mass mortality in both anurans and urodeles worldwide; however, the phylogenetic origin of FV3-like ranaviruses is not well established. In Asia, three FV3-like ranaviruses have been reported in farmed populations
[...] Read more.
Frog virus 3 (FV3) in the genus Ranavirus of the family Iridoviridae causes mass mortality in both anurans and urodeles worldwide; however, the phylogenetic origin of FV3-like ranaviruses is not well established. In Asia, three FV3-like ranaviruses have been reported in farmed populations of amphibians and reptiles. Here, we report the first case of endemic FV3-like ranavirus infections in the Korean clawed salamander Onychodactylus koreanus, caught in wild mountain streams in the Republic of Korea (ROK), through whole-genome sequencing and phylogenetic analysis. Two isolated FV3-like ranaviruses (Onychodactylus koreanus ranavirus, OKRV1 and 2) showed high similarity with the Rana grylio virus (RGV, 91.5%) and Rana nigromaculata ranavirus (RNRV, 92.2%) but relatively low similarity with the soft-shelled turtle iridovirus (STIV, 84.2%) in open reading frame (ORF) comparisons. OKRV1 and 2 formed a monophyletic clade with previously known Asian FV3-like ranaviruses, a sister group of the New World FV3-like ranavirus clade. Our results suggest that OKRV1 and 2 are FV3-like ranaviruses endemic to the ROK, and RGV and RNRV might also be endemic strains in China, unlike previous speculation. Our data have great implications for the study of the phylogeny and spreading routes of FV3-like ranaviruses and suggest the need for additional detection and analysis of FV3-like ranaviruses in wild populations in Asian countries.
Full article
(This article belongs to the Special Issue Identifying and Characterizing Viral Infections in Reptiles, Amphibians, Fish and Other Aquatic Species)
Open AccessArticle
Transcriptomic Investigation of the Virus Spectrum Carried by Midges in Border Areas of Yunnan Province
by
Lifen Yang, Weichen Wu, Sa Cai, Jing Wang, Guopeng Kuang, Weihong Yang, Juan Wang, Xi Han, Hong Pan, Mang Shi and Yun Feng
Viruses 2024, 16(5), 674; https://doi.org/10.3390/v16050674 - 25 Apr 2024
Abstract
Yunnan province in China shares its borders with three neighboring countries: Myanmar, Vietnam, and Laos. The region is characterized by a diverse climate and is known to be a suitable habitat for various arthropods, including midges which are notorious for transmitting diseases which
[...] Read more.
Yunnan province in China shares its borders with three neighboring countries: Myanmar, Vietnam, and Laos. The region is characterized by a diverse climate and is known to be a suitable habitat for various arthropods, including midges which are notorious for transmitting diseases which pose significant health burdens affecting both human and animal health. A total of 431,100 midges were collected from 15 different locations in the border region of Yunnan province from 2015 to 2020. These midges were divided into 37 groups according to the collection year and sampling site. These 37 groups of midges were then homogenized to extract nucleic acid. Metatranscriptomics were used to analyze their viromes. Based on the obtained cytochrome C oxidase I gene (COI) sequences, three genera were identified, including one species of Forcipomyia, one species of Dasyhelea, and twenty-five species of Culicoides. We identified a total of 3199 viruses in five orders and 12 families, including 1305 single-stranded positive-stranded RNA viruses (+ssRNA) in two orders and seven families, 175 single-stranded negative-stranded RNA viruses (−ssRNA) in two orders and one family, and 1719 double-stranded RNA viruses in five families. Six arboviruses of economic importance were identified, namely Banna virus (BAV), Japanese encephalitis virus (JEV), Akabane virus (AKV), Bluetongue virus (BTV), Tibetan circovirus (TIBOV), and Epizootic hemorrhagic disease virus (EHDV), all of which are capable, to varying extents, of causing disease in humans and/or animals. The survey sites in this study basically covered the current distribution area of midges in Yunnan province, which helps to predict the geographic expansion of midge species. The complexity and diversity of the viral spectrum carried by midges identified in the study calls for more in-depth research, which can be utilized to monitor arthropod vectors and to predict the emergence and spread of zoonoses and animal epidemics, which is of great significance for the control of vector-borne diseases.
Full article
(This article belongs to the Special Issue Vectors for Insect Viruses)
►▼
Show Figures
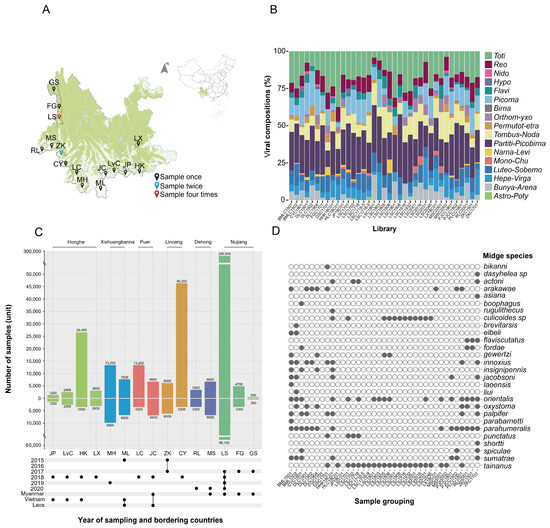
Figure 1
Open AccessReview
Towards Understanding and Identification of Human Viral Co-Infections
by
Hui Wu, Hang-Yu Zhou, Heng Zheng and Aiping Wu
Viruses 2024, 16(5), 673; https://doi.org/10.3390/v16050673 - 25 Apr 2024
Abstract
Viral co-infections, in which a host is infected with multiple viruses simultaneously, are common in the human population. Human viral co-infections can lead to complex interactions between the viruses and the host immune system, affecting the clinical outcome and posing challenges for treatment.
[...] Read more.
Viral co-infections, in which a host is infected with multiple viruses simultaneously, are common in the human population. Human viral co-infections can lead to complex interactions between the viruses and the host immune system, affecting the clinical outcome and posing challenges for treatment. Understanding the types, mechanisms, impacts, and identification methods of human viral co-infections is crucial for the prevention and control of viral diseases. In this review, we first introduce the significance of studying human viral co-infections and summarize the current research progress and gaps in this field. We then classify human viral co-infections into four types based on the pathogenic properties and species of the viruses involved. Next, we discuss the molecular mechanisms of viral co-infections, focusing on virus–virus interactions, host immune responses, and clinical manifestations. We also summarize the experimental and computational methods for the identification of viral co-infections, emphasizing the latest advances in high-throughput sequencing and bioinformatics approaches. Finally, we highlight the challenges and future directions in human viral co-infection research, aiming to provide new insights and strategies for the prevention, control, diagnosis, and treatment of viral diseases. This review provides a comprehensive overview of the current knowledge and future perspectives on human viral co-infections and underscores the need for interdisciplinary collaboration to address this complex and important topic.
Full article
(This article belongs to the Section Human Virology and Viral Diseases)
►▼
Show Figures
Figure 1
Open AccessArticle
Longitudinal Dynamics of Immune Response in Occupational Populations Post COVID-19 Infection in the Changning District of Shanghai, China
by
Li Li, Fengge Wang, Xiaoding He, Tingting Pei, Jiani Lu, Zhan Zhang, Ping Zhao, Jiayu Xue, Lin Zhu, Xinxin Chen, Zijie Yan, Yihan Lu and Jianlin Zhuang
Viruses 2024, 16(5), 672; https://doi.org/10.3390/v16050672 - 25 Apr 2024
Abstract
Monitoring the long-term changes in antibody and cellular immunity following Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) infection is crucial for understanding immune mechanisms that prevent reinfection. In March 2023, we recruited 167 participants from the Changning District, Shanghai, China. A subset of
[...] Read more.
Monitoring the long-term changes in antibody and cellular immunity following Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) infection is crucial for understanding immune mechanisms that prevent reinfection. In March 2023, we recruited 167 participants from the Changning District, Shanghai, China. A subset of 66 participants that were infected between November 2022 and January 2023 was selected for longitudinal follow-up. The study aimed to investigate the dynamics of the immune response, including neutralizing antibodies (NAbs), anti-spike (S)-immunoglobulin G (IgG), anti-S-IgM, and lymphocyte profiles, by analyzing peripheral blood samples collected three to seven months post infection. A gradual decrease in NAbs and IgG levels were observed from three to seven months post infection. No significant differences in NAbs and IgG titers were found across various demographics, including age, sex, occupation, and symptomatic presentation, across five follow-up assessments. Additionally, a strong correlation between NAbs and IgG levels was identified. Lymphocyte profiles showed a slight change at five months but had returned to baseline levels by seven months post infection. Notably, healthcare workers exhibited lower B-cell levels compared to police officers. Our study demonstrated that the immune response to SARS-CoV-2 infection persisted for at least seven months. Similar patterns in the dynamics of antibody responses and cellular immunity were observed throughout this period.
Full article
(This article belongs to the Section Coronaviruses)
►▼
Show Figures
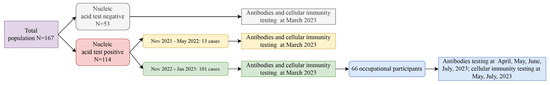
Figure 1

Journal Menu
► ▼ Journal Menu-
- Viruses Home
- Aims & Scope
- Editorial Board
- Reviewer Board
- Topical Advisory Panel
- Instructions for Authors
- Special Issues
- Topics
- Sections & Collections
- Article Processing Charge
- Indexing & Archiving
- Editor’s Choice Articles
- Most Cited & Viewed
- Journal Statistics
- Journal History
- Journal Awards
- Society Collaborations
- Conferences
- Editorial Office
Journal Browser
► ▼ Journal BrowserHighly Accessed Articles
Latest Books
E-Mail Alert
News
Topics
Topic in
Diseases, Infectious Disease Reports, Pathogens, Viruses, TropicalMed
Human Monkeypox Research
Topic Editors: Shailendra K. Saxena, Ahmed Sayed Abdel-MoneimDeadline: 30 June 2024
Topic in
Biomedicines, JCM, Pathogens, Vaccines, Viruses
Discovery and Development of Monkeypox Disease Treatments
Topic Editors: Mohd Imran, Ali A. RabaanDeadline: 31 August 2024
Topic in
Brain Sciences, Clinics and Practice, COVID, Life, Vaccines, Viruses
Multifaceted Efforts from Basic Research to Clinical Practice in Controlling COVID-19 Disease
Topic Editors: Yih-Horng Shiao, Rashi OjhaDeadline: 30 September 2024

Conferences
Special Issues
Special Issue in
Viruses
Oncolytic Viruses as Immunotherapeutic Agents
Guest Editor: Nadine Van MontfoortDeadline: 1 May 2024
Special Issue in
Viruses
Animal Coronaviruses: Infection, Prevention, and Antivirals
Guest Editor: Tomomi TakanoDeadline: 20 May 2024
Special Issue in
Viruses
Bacteriophages and Biofilms 2.0
Guest Editors: Zuzanna Drulis-Kawa, Tomasz OlszakDeadline: 31 May 2024
Special Issue in
Viruses
The Inflammasomes - Key Players in Antiviral Response
Guest Editors: Dong-Yan Jin, Tsan Sam XiaoDeadline: 15 June 2024
Topical Collections
Topical Collection in
Viruses
Poxviruses
Collection Editors: Giliane de Souza Trindade, Galileu Barbosa Costa, Flavio Guimaraes da Fonseca
Topical Collection in
Viruses
Phage Therapy
Collection Editors: Nina Chanishvili, Jean-Paul Pirnay, Mikael Skurnik
Topical Collection in
Viruses
Coronaviruses
Collection Editors: Luis Martinez-Sobrido, Fernando Almazan Toral
Topical Collection in
Viruses
SARS-CoV-2 and COVID-19
Collection Editors: Luis Martinez-Sobrido, Fernando Almazan Toral








