Journal Description
Antibiotics
Antibiotics
is an international, peer-reviewed, open access journal on all aspects of antibiotics, published monthly online by MDPI.
- Open Access— free for readers, with article processing charges (APC) paid by authors or their institutions.
- High Visibility: indexed within Scopus, SCIE (Web of Science), PubMed, PMC, Embase, CAPlus / SciFinder, and other databases.
- Journal Rank: JCR - Q1 (Pharmacology & Pharmacy) / CiteScore - Q1 (General Pharmacology, Toxicology and Pharmaceutics)
- Rapid Publication: manuscripts are peer-reviewed and a first decision is provided to authors approximately 13.7 days after submission; acceptance to publication is undertaken in 2.5 days (median values for papers published in this journal in the second half of 2023).
- Recognition of Reviewers: reviewers who provide timely, thorough peer-review reports receive vouchers entitling them to a discount on the APC of their next publication in any MDPI journal, in appreciation of the work done.
Impact Factor:
4.8 (2022);
5-Year Impact Factor:
4.9 (2022)
Latest Articles
Microbiology of Prosthetic Joint Infections: A Retrospective Study of an Italian Orthopaedic Hospital
Antibiotics 2024, 13(5), 399; https://doi.org/10.3390/antibiotics13050399 - 26 Apr 2024
Abstract
The most frequent cause of periprosthetic infections (PJIs) is intraoperative contamination; hence, antibiotic prophylaxis plays a crucial role in prevention. Modifications to standard prophylaxis can be considered if there is a high incidence of microorganisms resistant to current protocols. To date, very few
[...] Read more.
The most frequent cause of periprosthetic infections (PJIs) is intraoperative contamination; hence, antibiotic prophylaxis plays a crucial role in prevention. Modifications to standard prophylaxis can be considered if there is a high incidence of microorganisms resistant to current protocols. To date, very few studies regarding microbial etiology have been published in Italy. In this single-center, retrospective study conducted at IRCCS Ospedale Galeazzi-Sant’Ambrogio in Milan, we analyzed hip, knee, and shoulder PJIs in patients undergoing first implantation between 1 January 17 and 31 December 2021. The primary aim was to derive a local microbiological case history. The secondary aim was to evaluate the adequacy of preoperative antibiotic prophylaxis in relation to the identified bacteria. A total of 57 PJIs and 65 pathogens were identified: 16 S. aureus, 15 S. epidermidis, and 10 other coagulase-negative staphylococci (CoNS), which accounted for 63% of the isolations. A total of 86.7% of S. epidermidis were methicillin-resistant (MRSE). In line with other case reports, we found a predominance of staphylococcal infections, with a lower percentage of MRSA than the Italian average, while we found a high percentage of MRSE. We estimated that 44.6% of the bacteria isolated were resistant to cefazolin, our standard prophylaxis. These PJIs could be prevented by using glycopeptide alone or in combination with cefazolin, but the literature reports conflicting results regarding the adequacy of such prophylaxis. In conclusion, our study showed that in our local hospital, our standard antibiotic prophylaxis is ineffective for almost half of the cases, highlighting the importance of defining specific antibiotic guidelines based on the local bacterial prevalence of each institution.
Full article
(This article belongs to the Special Issue Antimicrobial Therapeutics for Bone and Periprosthetic Joint Infection)
Open AccessArticle
Discovery of Antimicrobial Agents Based on Structural and Functional Study of the Klebsiella pneumoniae MazEF Toxin–Antitoxin System
by
Chenglong Jin, Sung-Min Kang, Do-Hee Kim, Yuno Lee and Bong-Jin Lee
Antibiotics 2024, 13(5), 398; https://doi.org/10.3390/antibiotics13050398 - 26 Apr 2024
Abstract
Klebsiella pneumoniae causes severe human diseases, but its resistance to current antibiotics is increasing. Therefore, new antibiotics to eradicate K. pneumoniae are urgently needed. Bacterial toxin–antitoxin (TA) systems are strongly correlated with physiological processes in pathogenic bacteria, such as growth arrest, survival, and
[...] Read more.
Klebsiella pneumoniae causes severe human diseases, but its resistance to current antibiotics is increasing. Therefore, new antibiotics to eradicate K. pneumoniae are urgently needed. Bacterial toxin–antitoxin (TA) systems are strongly correlated with physiological processes in pathogenic bacteria, such as growth arrest, survival, and apoptosis. By using structural information, we could design the peptides and small-molecule compounds that can disrupt the binding between K. pneumoniae MazE and MazF, which release free MazF toxin. Because the MazEF system is closely implicated in programmed cell death, artificial activation of MazF can promote cell death of K. pneumoniae. The effectiveness of a discovered small-molecule compound in bacterial cell killing was confirmed through flow cytometry analysis. Our findings can contribute to understanding the bacterial MazEF TA system and developing antimicrobial agents for treating drug-resistant K. pneumoniae.
Full article
Open AccessArticle
Associations between Polycystic Ovary Syndrome (PCOS) and Antibiotic Use: Results from the UAEHFS
by
Nirmin F. Juber, Abdishakur Abdulle, Amar Ahmad, Fatme AlAnouti, Tom Loney, Youssef Idaghdour, Yvonne Valles and Raghib Ali
Antibiotics 2024, 13(5), 397; https://doi.org/10.3390/antibiotics13050397 - 26 Apr 2024
Abstract
Women with polycystic ovary syndrome (PCOS) have a higher susceptibility to infections compared to those without PCOS. Studies evaluating antibiotic use based on PCOS status are scarce. Therefore, we aimed to (i) assess the associations between self-reported PCOS and antibiotic use, and (ii)
[...] Read more.
Women with polycystic ovary syndrome (PCOS) have a higher susceptibility to infections compared to those without PCOS. Studies evaluating antibiotic use based on PCOS status are scarce. Therefore, we aimed to (i) assess the associations between self-reported PCOS and antibiotic use, and (ii) whether PCOS treatment and the age at PCOS diagnosis modified the associations above. This cross-sectional analysis used the United Arab Emirates Healthy Future Study (UAEHFS) conducted from February 2016 to March 2023 involving 2063 Emirati women aged 18–62 years. We performed ordinal logistic regressions under unadjusted and demographic-health-characteristic-adjusted models to obtain the odds ratios (ORs) and 95% confidence intervals (CIs) to analyze PCOS and antibiotic use. Subgroup analyses were performed by treatment status and age at diagnosis. We found that women with PCOS were 55% more likely to frequently take a course of antibiotics in the past year (aOR 1.55; 95% CI 1.26–1.90). Similar likelihoods were also found among those being treated for PCOS and those without treatment but with a PCOS diagnosis at ≤25 years. Our study suggests that PCOS was associated with an increased use of antibiotics among Emirati women. Understanding the frequent antibiotic use susceptibility among those with PCOS may improve antibiotic use surveillance and promote antibiotic stewardship in these at-risk individuals.
Full article
(This article belongs to the Special Issue Antimicrobial Prescribing, Population Use and Resistance, Impact in Global Health)
►▼
Show Figures
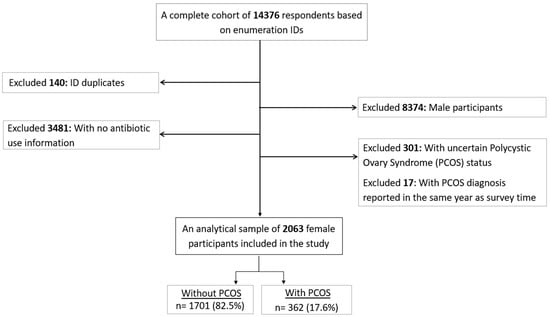
Figure 1
Open AccessArticle
Exploring the Molecular Mechanisms of Macrolide Resistance in Laboratory Mutant Helicobacter pylori
by
Meltem Ayaş, Sinem Oktem-Okullu, Orhan Özcan, Tanıl Kocagöz and Yeşim Gürol
Antibiotics 2024, 13(5), 396; https://doi.org/10.3390/antibiotics13050396 - 26 Apr 2024
Abstract
Resistance to clarithromycin, a macrolide antibiotic used in the first-line treatment of Helicobacter pylori infection, is the most important cause of treatment failure. Although most cases of clarithromycin resistance in H. pylori are associated with point mutations in 23S ribosomal RNA (rRNA), the
[...] Read more.
Resistance to clarithromycin, a macrolide antibiotic used in the first-line treatment of Helicobacter pylori infection, is the most important cause of treatment failure. Although most cases of clarithromycin resistance in H. pylori are associated with point mutations in 23S ribosomal RNA (rRNA), the relationships of other mutations with resistance remain unclear. We examined possible new macrolide resistance mechanisms in resistant strains using next-generation sequencing. Two resistant strains were obtained from clarithromycin-susceptible H. pylori following exposure to low clarithromycin concentrations using the agar dilution method. Sanger sequencing and whole-genome sequencing were performed to detect resistance-related mutations. Both strains carried the A2142G mutation in 23S rRNA. Candidate mutations (T1495A, T1494A, T1490A, T1476A, and G1472T) for clarithromycin resistance were detected in the Mutant-1 strain. Furthermore, a novel mutation in the gene encoding for the sulfite exporter TauE/SafE family protein was considered to be linked to clarithromycin resistance or cross-resistance, being identified as a target for further investigations. In the Mutant-2 strain, a novel mutation in the gene that encodes DUF874 family protein that can be considered as relevant with antibiotic resistance was detected. These mutations were revealed in the H. pylori genome for the first time, emphasizing their potential as targets for advanced studies.
Full article
(This article belongs to the Special Issue Helicobacter pylori Infection and Antibiotic Resistance: The Emerging Eradication Strategies)
►▼
Show Figures

Figure 1
Open AccessArticle
Low Oxygen Concentration Reduces Neisseria gonorrhoeae Susceptibility to Resazurin
by
Justin Rice, Jordan Gibson, Emily Young, Kendall Souder, Kailee Cunningham and Deanna M. Schmitt
Antibiotics 2024, 13(5), 395; https://doi.org/10.3390/antibiotics13050395 - 26 Apr 2024
Abstract
Neisseria gonorrhoeae has developed resistance to every antibiotic currently approved for the treatment of gonorrhea, prompting the development of new therapies. The phenoxazine dye resazurin exhibits robust antimicrobial activity against N. gonorrhoeae in vitro but fails to limit vaginal colonization by N. gonorrhoeae
[...] Read more.
Neisseria gonorrhoeae has developed resistance to every antibiotic currently approved for the treatment of gonorrhea, prompting the development of new therapies. The phenoxazine dye resazurin exhibits robust antimicrobial activity against N. gonorrhoeae in vitro but fails to limit vaginal colonization by N. gonorrhoeae in a mouse model. The lack of in vivo efficacy may be due to oxygen limitation as in vitro susceptibility assays with resazurin are conducted under atmospheric oxygen while a microaerophilic environment is present in the vagina. Here, we utilized broth microdilution assays to determine the susceptibility of N. gonorrhoeae to resazurin under low and atmospheric oxygen conditions. The minimal inhibitory concentration of resazurin for multiple N. gonorrhoeae clinical isolates was significantly higher under low oxygen. This effect was specific to resazurin as N. gonorrhoeae was equally susceptible to other antibiotics under low and atmospheric oxygen conditions. The reduced susceptibility of N. gonorrhoeae to resazurin under low oxygen was largely attributed to reduced oxidative stress, as the addition of antioxidants under atmospheric oxygen mimicked the reduced susceptibility to resazurin observed under low oxygen. Together, these data suggest oxygen concentration is an important factor to consider when evaluating the efficacy of new antibiotics against N. gonorrhoeae in vitro.
Full article
(This article belongs to the Special Issue Novel Approaches for the Treatment of Antimicrobial-Resistant Pathogens)
►▼
Show Figures
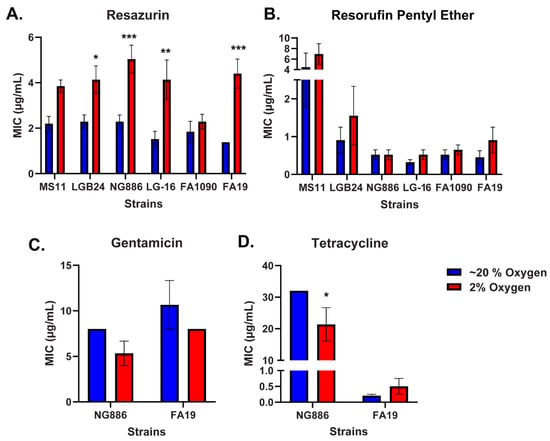
Figure 1
Open AccessCommunication
Detecting Class 1 Integrons and Their Variable Regions in Escherichia coli Whole-Genome Sequences Reported from Andean Community Countries
by
María Nicole Solis, Karen Loaiza, Lilibeth Torres-Elizalde, Ivan Mina, Miroslava Anna Šefcová and Marco Larrea-Álvarez
Antibiotics 2024, 13(5), 394; https://doi.org/10.3390/antibiotics13050394 - 25 Apr 2024
Abstract
Various genetic elements, including integrons, are known to contribute to the development of antimicrobial resistance. Class 1 integrons have been identified in E. coli isolates and are associated with multidrug resistance in countries of the Andean Community. However, detailed information on the gene
[...] Read more.
Various genetic elements, including integrons, are known to contribute to the development of antimicrobial resistance. Class 1 integrons have been identified in E. coli isolates and are associated with multidrug resistance in countries of the Andean Community. However, detailed information on the gene cassettes located on the variable regions of integrons is lacking. Here, we investigated the presence and diversity of class 1 integrons, using an in silico approach, in 2533 whole-genome sequences obtained from EnteroBase. IntFinder v1.0 revealed that almost one-third of isolates contained these platforms. Integron-bearing isolates were associated with environmental, food, human, and animal origins reported from all countries under scrutiny. Moreover, they were identified in clones known for their pathogenicity or multidrug resistance. Integrons carried cassettes associated with aminoglycoside (aadA), trimethoprim (dfrA), cephalosporin (blaOXA; blaDHA), and fluoroquinolone (aac(6′)-Ib-cr; qnrB) resistance. These platforms showed higher diversity and larger numbers than previously reported. Moreover, integrons carrying more than three cassettes in their variable regions were determined. Monitoring the prevalence and diversity of genetic elements is necessary for recognizing emergent patterns of resistance in pathogenic bacteria, especially in countries where various factors are recognized to favor the selection of resistant microorganisms.
Full article
(This article belongs to the Special Issue Genomic Analysis of Antimicrobial Drug-Resistant Bacteria)
Open AccessArticle
New Insights into the Biological Functions of Essential TsaB/YeaZ Protein in Staphylococcus aureus
by
Haiyong Guo, Ting Lei, Junshu Yang, Yue Wang, Yifan Wang and Yinduo Ji
Antibiotics 2024, 13(5), 393; https://doi.org/10.3390/antibiotics13050393 - 25 Apr 2024
Abstract
TsaB/YeaZ represents a promising target for novel antibacterial agents due to its indispensable role in bacterial survival, high conservation within bacterial species, and absence of eukaryotic homologs. Previous studies have elucidated the role of the essential staphylococcal protein, TsaB/YeaZ, in binding DNA to
[...] Read more.
TsaB/YeaZ represents a promising target for novel antibacterial agents due to its indispensable role in bacterial survival, high conservation within bacterial species, and absence of eukaryotic homologs. Previous studies have elucidated the role of the essential staphylococcal protein, TsaB/YeaZ, in binding DNA to mediate the transcription of the ilv-leu operon, responsible for encoding key enzymes involved in the biosynthesis of branched-chain amino acids—namely isoleucine, leucine, and valine (ILV). However, the regulation of ILV biosynthesis does not account for the essentiality of TsaB/YeaZ for bacterial growth. In this study, we investigated the impact of TsaB/YeaZ depletion on bacterial morphology and gene expression profiles using electron microscopy and deep transcriptomic analysis, respectively. Our results revealed significant alterations in bacterial size and surface smoothness upon TsaB/YeaZ depletion. Furthermore, we pinpointed specific genes and enriched biological pathways significantly affected by TsaB/YeaZ during the early and middle exponential phases and early stationary phases of growth. Crucially, our research uncovered a regulatory role for TsaB/YeaZ in bacterial autolysis. These discoveries offer fresh insights into the multifaceted biological functions of TsaB/YeaZ within S. aureus.
Full article
(This article belongs to the Special Issue Genomic Analysis of Antimicrobial Drug-Resistant Bacteria)
Open AccessReview
The Role of Gut Microbiota in the Etiopathogenesis of Multiple Chronic Diseases
by
Lara Pires, Ana M. González-Paramás, Sandrina A. Heleno and Ricardo C. Calhelha
Antibiotics 2024, 13(5), 392; https://doi.org/10.3390/antibiotics13050392 - 25 Apr 2024
Abstract
Chronic diseases (CD) may result from a combination of genetic factors, lifestyle and social behaviours, healthcare system influences, community factors, and environmental determinants of health. These risk factors frequently coexist and interact with one another. Ongoing research and a focus on personalized interventions
[...] Read more.
Chronic diseases (CD) may result from a combination of genetic factors, lifestyle and social behaviours, healthcare system influences, community factors, and environmental determinants of health. These risk factors frequently coexist and interact with one another. Ongoing research and a focus on personalized interventions are pivotal strategies for preventing and managing chronic disease outcomes. A wealth of literature suggests the potential involvement of gut microbiota in influencing host metabolism, thereby impacting various risk factors associated with chronic diseases. Dysbiosis, the perturbation of the composition and activity of the gut microbiota, is crucial in the etiopathogenesis of multiple CD. Recent studies indicate that specific microorganism-derived metabolites, including trimethylamine N-oxide, lipopolysaccharide and uremic toxins, contribute to subclinical inflammatory processes implicated in CD. Various factors, including diet, lifestyle, and medications, can alter the taxonomic species or abundance of gut microbiota. Researchers are currently dedicating efforts to understanding how the natural progression of microbiome development in humans affects health outcomes. Simultaneously, there is a focus on enhancing the understanding of microbiome–host molecular interactions. These endeavours ultimately aim to devise practical approaches for rehabilitating dysregulated human microbial ecosystems, intending to restore health and prevent diseases. This review investigates how the gut microbiome contributes to CD and explains ways to modulate it for managing or preventing chronic conditions.
Full article
Open AccessReview
Extended-Spectrum Beta-Lactamase-Producing Escherichia coli and Other Antimicrobial-Resistant Gram-Negative Pathogens Isolated from Bovine Mastitis: A One Health Perspective
by
Breno Luis Nery Garcia, Stéfani Thais Alves Dantas, Kristian da Silva Barbosa, Thatiane Mendes Mitsunaga, Alyssa Butters, Carlos Henrique Camargo and Diego Borin Nobrega
Antibiotics 2024, 13(5), 391; https://doi.org/10.3390/antibiotics13050391 - 25 Apr 2024
Abstract
Antimicrobial resistance (AMR) poses an imminent threat to global public health, driven in part by the widespread use of antimicrobials in both humans and animals. Within the dairy cattle industry, Gram-negative coliforms such as Escherichia coli and Klebsiella pneumoniae stand out as major
[...] Read more.
Antimicrobial resistance (AMR) poses an imminent threat to global public health, driven in part by the widespread use of antimicrobials in both humans and animals. Within the dairy cattle industry, Gram-negative coliforms such as Escherichia coli and Klebsiella pneumoniae stand out as major causative agents of clinical mastitis. These same bacterial species are frequently associated with severe infections in humans, including bloodstream and urinary tract infections, and contribute significantly to the alarming surge in antimicrobial-resistant bacterial infections worldwide. Additionally, mastitis-causing coliforms often carry AMR genes akin to those found in hospital-acquired strains, notably the extended-spectrum beta-lactamase genes. This raises concerns regarding the potential transmission of resistant bacteria and AMR from mastitis cases in dairy cattle to humans. In this narrative review, we explore the distinctive characteristics of antimicrobial-resistant E. coli and Klebsiella spp. strains implicated in clinical mastitis and human infections. We focus on the molecular mechanisms underlying AMR in these bacterial populations and critically evaluate the potential for interspecies transmission. Despite some degree of similarity observed in sequence types and mobile genetic elements between strains found in humans and cows, the existing literature does not provide conclusive evidence to assert that coliforms responsible for mastitis in cows pose a direct threat to human health. Finally, we also scrutinize the existing literature, identifying gaps and limitations, and propose avenues for future research to address these pressing challenges comprehensively.
Full article
(This article belongs to the Special Issue Antimicrobial Resistance of Pathogens Isolated from Bovine Mastitis)
Open AccessArticle
Pseudomonas aeruginosa Infections in Patients with Severe COVID-19 in Intensive Care Units: A Retrospective Study
by
Alexandre Baudet, Marie Regad, Sébastien Gibot, Élodie Conrath, Julie Lizon, Béatrice Demoré and Arnaud Florentin
Antibiotics 2024, 13(5), 390; https://doi.org/10.3390/antibiotics13050390 - 25 Apr 2024
Abstract
Patients hospitalized in ICUs with severe COVID-19 are at risk for developing hospital-acquired infections, especially infections caused by Pseudomonas aeruginosa. We aimed to describe the evolution of P. aeruginosa infections in ICUs at CHRU-Nancy (France) in patients with severe COVID-19 during the
[...] Read more.
Patients hospitalized in ICUs with severe COVID-19 are at risk for developing hospital-acquired infections, especially infections caused by Pseudomonas aeruginosa. We aimed to describe the evolution of P. aeruginosa infections in ICUs at CHRU-Nancy (France) in patients with severe COVID-19 during the three initial waves of COVID-19. The second aims were to analyze P. aeruginosa resistance and to describe the antibiotic treatments. We conducted a retrospective cohort study among adult patients who were hospitalized for acute respiratory distress syndrome due to COVID-19 and who developed a hospital-acquired infection caused by P. aeruginosa during their ICU stay. Among the 51 patients included, most were male (90%) with comorbidities (77%), and the first identification of P. aeruginosa infection occurred after a median ICU stay of 11 days. Several patients acquired infections with MDR (27%) and XDR (8%) P. aeruginosa strains. The agents that strains most commonly exhibited resistance to were penicillin + β-lactamase inhibitors (59%), cephalosporins (42%), monobactams (32%), and carbapenems (27%). Probabilistic antibiotic treatment was prescribed for 49 patients (96%) and was subsequently adapted for 51% of patients after antibiogram and for 33% of patients after noncompliant antibiotic plasma concentration. Hospital-acquired infection is a common and life-threatening complication in critically ill patients. Efforts to minimize the occurrence and improve the treatment of such infections, including infections caused by resistant strains, must be pursued.
Full article
(This article belongs to the Special Issue Successful Antimicrobial Stewardship Approaches to Address Nosocomial Infections)
►▼
Show Figures
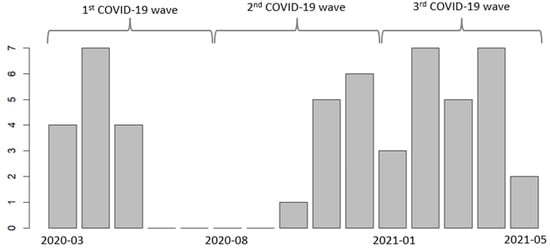
Figure 1
Open AccessArticle
Long-Term Co-Circulation of Host-Specialist and Host-Generalist Lineages of Group B Streptococcus in Brazilian Dairy Cattle with Heterogeneous Antimicrobial Resistance Profiles
by
Laura Maria Andrade de Oliveira, Leandro Correia Simões, Chiara Crestani, Natália Silva Costa, José Carlos de Figueiredo Pantoja, Renata Fernandes Rabello, Lucia Martins Teixeira, Uzma Basit Khan, Stephen Bentley, Dorota Jamrozy, Tatiana de Castro Abreu Pinto and Ruth N. Zadoks
Antibiotics 2024, 13(5), 389; https://doi.org/10.3390/antibiotics13050389 - 25 Apr 2024
Abstract
Group B Streptococcus (GBS) is a major cause of contagious bovine mastitis (CBM) in Brazil. The GBS population is composed of host-generalist and host-specialist lineages, which may differ in antimicrobial resistance (AMR) and zoonotic potential, and the surveillance of bovine GBS is crucial
[...] Read more.
Group B Streptococcus (GBS) is a major cause of contagious bovine mastitis (CBM) in Brazil. The GBS population is composed of host-generalist and host-specialist lineages, which may differ in antimicrobial resistance (AMR) and zoonotic potential, and the surveillance of bovine GBS is crucial to developing effective CBM control and prevention measures. Here, we investigated bovine GBS isolates (n = 156) collected in Brazil between 1987 and 2021 using phenotypic testing and whole-genome sequencing to uncover the molecular epidemiology of bovine GBS. Clonal complex (CC) 61/67 was the predominant clade in the 20th century; however, it was replaced by CC91, with which it shares a most common recent ancestor, in the 21st century, despite the higher prevalence of AMR in CC61/67 than in CC91, and high selection pressure for AMR from indiscriminate antimicrobial use in the Brazilian dairy industry. CC103 also emerged as a dominant CC in the 21st century, and a considerable proportion of herds had two or more GBS strains, suggesting poor biosecurity and within-herd evolution due to the chronic nature of CBM problems. The majority of bovine GBS belonged to serotype Ia or III, which was strongly correlated with CCs. Ninety-three isolates were resistant to tetracycline (≥8 μg/mL; tetO = 57, tetM = 34 or both = 2) and forty-four were resistant to erythromycin (2.0 to >4 μg/mL; ermA = 1, ermB = 38, mechanism unidentified n = 5). Only three isolates were non-susceptible to penicillin (≥8.0 μg/mL), providing opportunities for improved antimicrobial stewardship through the use of narrow-spectrum antimicrobials for the treatment of dairy cattle. The common bovine GBS clades detected in this study have rarely been reported in humans, suggesting limited risk of interspecies transmission of GBS in Brazil. This study provides new data to support improvements to CBM and AMR control, bovine GBS vaccine design, and the management of public health risks posed by bovine GBS in Brazil.
Full article
(This article belongs to the Special Issue Antimicrobial Resistance of Pathogens Isolated from Bovine Mastitis)
►▼
Show Figures
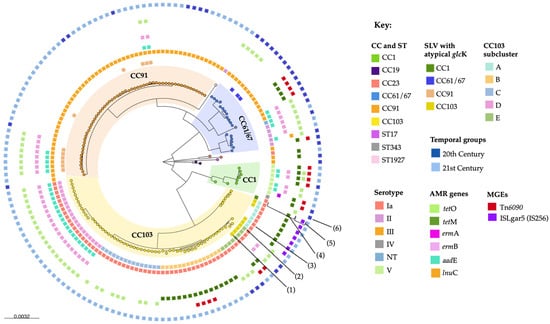
Figure 1
Open AccessArticle
Factors Associated with Prolonged Antibiotic Therapy in Neonates with Suspected Early-Onset Sepsis
by
Bo M. van der Weijden, Jolien R. van Dorth, Niek B. Achten and Frans B. Plötz
Antibiotics 2024, 13(5), 388; https://doi.org/10.3390/antibiotics13050388 - 25 Apr 2024
Abstract
Early-onset sepsis (EOS) is a rare but profoundly serious bacterial infection. Neonates at risk of EOS are often treated with antibiotics. The start of empiric antibiotic therapy can successfully be reduced by the implementation of the EOS calculator. However, once started, antibiotic therapy
[...] Read more.
Early-onset sepsis (EOS) is a rare but profoundly serious bacterial infection. Neonates at risk of EOS are often treated with antibiotics. The start of empiric antibiotic therapy can successfully be reduced by the implementation of the EOS calculator. However, once started, antibiotic therapy is often continued despite a negative blood culture. To decrease the burden of antibiotic therapy, it is necessary to know whether the clinician’s reasons are based on objective factors. Therefore, we performed a retrospective single-centre cohort study to identify the factors associated with prolongation of antibiotic therapy in neonates with suspected EOS but a negative blood culture. Maternal, clinical, and laboratory data of neonates with a gestational age of ≥32 weeks, admitted between January 2019 and June 2021, were collected. Among neonates with a negative blood culture, we compared neonates with prolonged (≥3 days) to neonates with discontinued (<3 days) antibiotic therapy. The clinician’s reported reasons for prolonging therapy were explored. Blood cultures were positive in 4/146 (2.7%), negative in 131/146 (89.7%), and not obtained in 11/146 (7.5%) of the neonates. The incidence of EOS was 0.7 per 1000 neonates. Of the 131 neonates with a negative blood culture, 47 neonates (35.9%) received prolonged antibiotic therapy. In the prolonged group, the mean gestational age was higher (38.9 versus 36.8 weeks), and spontaneous preterm birth was less prevalent (21.3% versus 53.6%). Prolonged treatment was associated with late onset of respiratory distress, respiratory rate, hypoxia, apnoea and bradycardia, pale appearance, behavioural change, and elevated CRP levels. The most reported reasons were clinical appearance (38.3%), elevated CRP levels (36.2%), and skin colour (10.6%). Prolonging empiric antibiotic therapy despite a negative blood culture is common in suspected EOS. Clinical signs associated with prolongation are uncommon and the reported reasons for prolongation contain subjective assessments and arbitrary interpretations that are not supported by the guideline recommendations as arguments for prolonged therapy.
Full article
(This article belongs to the Special Issue Neonatal Infections: Epidemiology, Diagnostics and Antibiotic Therapeutics)
Open AccessCase Report
Cerebral Infectious Opportunistic Lesions in a Patient with Acute Myeloid Leukaemia: The Challenge of Diagnosis and Clinical Management
by
Gabriele Cavazza, Cristina Motto, Caroline Regna-Gladin, Giovanna Travi, Elisa Di Gennaro, Francesco Peracchi, Bianca Monti, Nicolò Corti, Rosa Greco, Periana Minga, Marta Riva, Sara Rimoldi, Marta Vecchi, Carlotta Rogati, Davide Motta, Annamaria Pazzi, Chiara Vismara, Laura Bandiera, Fulvio Crippa, Valentina Mancini, Maria Sessa, Chiara Oltolini, Roberto Cairoli and Massimo Puotiadd
Show full author list
remove
Hide full author list
Antibiotics 2024, 13(5), 387; https://doi.org/10.3390/antibiotics13050387 - 24 Apr 2024
Abstract
Central nervous system (CNS) lesions, especially invasive fungal diseases (IFDs), in immunocompromised patients pose a great challenge in diagnosis and treatment. We report the case of a 48-year-old man with acute myeloid leukaemia and probable pulmonary aspergillosis, who developed hyposthenia of the left
[...] Read more.
Central nervous system (CNS) lesions, especially invasive fungal diseases (IFDs), in immunocompromised patients pose a great challenge in diagnosis and treatment. We report the case of a 48-year-old man with acute myeloid leukaemia and probable pulmonary aspergillosis, who developed hyposthenia of the left upper limb, after achieving leukaemia remission and while on voriconazole. Magnetic resonance imaging (MRI) showed oedematous CNS lesions with a haemorrhagic component in the right hemisphere with lepto-meningitis. After 2 weeks of antibiotics and amphotericin-B, brain biopsy revealed chronic inflammation with abscess and necrosis, while cultures were negative. Clinical recovery was attained, he was discharged on isavuconazole and allogeneic transplant was postponed, introducing azacitidine as a maintenance therapy. After initial improvement, MRI worsened; brain biopsy was repeated, showing similar histology; and 16S metagenomics sequencing analysis was positive (Veilonella, Pseudomonas). Despite 1 month of meropenem, MRI did not improve. The computer tomography and PET scan excluded extra-cranial infectious–inflammatory sites, and auto-immune genesis (sarcoidosis, histiocytosis, CNS vasculitis) was deemed unlikely due to the histological findings and unilateral lesions. We hypothesised possible IFD with peri-lesion inflammation and methyl-prednisolone was successfully introduced. Steroid tapering is ongoing and isavuconazole discontinuation is planned with close follow-up. In conclusion, the management of CNS complications in immunocompromised patients needs an interdisciplinary approach.
Full article
(This article belongs to the Special Issue Antimicrobial Treatment and Management of Central Nervous System Infections)
►▼
Show Figures
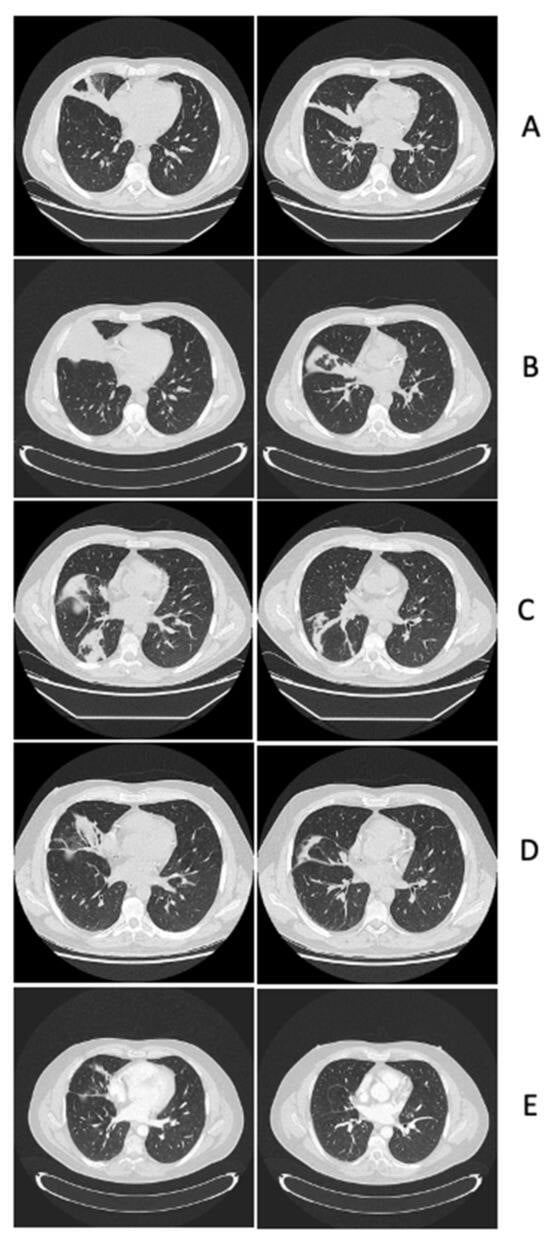
Figure 1
Open AccessArticle
Efficacy of Tamoxifen Metabolites in Combination with Colistin and Tigecycline in Experimental Murine Models of Escherichia coli and Acinetobacter baumannii
by
Soraya Herrera-Espejo, Andrea Vila-Domínguez, Tania Cebrero-Cangueiro, Younes Smani, Jerónimo Pachón, Manuel E. Jiménez-Mejías and María E. Pachón-Ibáñez
Antibiotics 2024, 13(5), 386; https://doi.org/10.3390/antibiotics13050386 - 24 Apr 2024
Abstract
This study aimed to evaluate the potential of tamoxifen and N-desmethyltamoxifen metabolites as therapeutic agents against multidrug-resistant Escherichia coli and Acinetobacter baumannii, using a repurposing approach to shorten the time required to obtain a new effective treatment against multidrug-resistant bacterial infections. Characterisation and
[...] Read more.
This study aimed to evaluate the potential of tamoxifen and N-desmethyltamoxifen metabolites as therapeutic agents against multidrug-resistant Escherichia coli and Acinetobacter baumannii, using a repurposing approach to shorten the time required to obtain a new effective treatment against multidrug-resistant bacterial infections. Characterisation and virulence studies were conducted on E. coli (colistin-susceptible C1-7-LE and colistin-resistant MCR-1+) and A. baumannii (tigecycline-susceptible Ab#9 and tigecycline-resistant Ab#186) strains. The efficacy of the metabolite mix (33.3% each) and N-desmethyltamoxifen in combination with colistimethate sodium (CMS) or tigecycline was evaluated in experimental models in mice. In the pneumonia model, N-desmethyltamoxifen exhibited significant efficacy against Ab#9 and both E. coli strains, especially E. coli MCR-1+ (−2.86 log10 CFU/g lungs, −5.88 log10 CFU/mL blood, and −50% mortality), and against the Ab#186 strain when combined with CMS (−2.27 log10 CFU/g lungs, −2.73 log10 CFU/mL blood, and −40% mortality) or tigecycline (−3.27 log10 CFU/g lungs, −4.95 log10 CFU/mL blood, and −50% mortality). Moreover, the metabolite mix in combination with both antibiotics decreased the bacterial concentrations in the lungs and blood for both A. baumannii strains. In the sepsis model, the significant efficacy of the metabolite mix was restricted to the colistin-susceptible E. coli C1-7-LE strain (−3.32 log10 CFU/g lung, −6.06 log10 CFU/mL blood, and −79% mortality). N-desmethyltamoxifen could be a new therapeutic option in combination with CMS or tigecycline for combating multidrug-resistant GNB, specifically A. baumannii.
Full article
(This article belongs to the Special Issue Advancing the Discovery and Development of New Antibiotics through Drug Repurposing)
Open AccessArticle
Use of the Naturally Occurring Bacteriophage Grouping Model for the Design of Potent Therapeutic Cocktails
by
Tea Glonti, Michael Goossens, Christel Cochez, Sabrina Green, Sayali Gorivale, Jeroen Wagemans, Rob Lavigne and Jean-Paul Pirnay
Antibiotics 2024, 13(5), 385; https://doi.org/10.3390/antibiotics13050385 - 24 Apr 2024
Abstract
The specificity of phages and their ability to evolve and overcome bacterial resistance make them potentially useful as adjuncts in the treatment of antibiotic-resistant bacterial infections. The goal of this study was to mimic a natural grouping of phages of interest and to
[...] Read more.
The specificity of phages and their ability to evolve and overcome bacterial resistance make them potentially useful as adjuncts in the treatment of antibiotic-resistant bacterial infections. The goal of this study was to mimic a natural grouping of phages of interest and to evaluate the nature of their proliferation dynamics with bacteria. We have, for the first time, transferred naturally occurring phage groups directly from their sources of isolation to in vitro and identified 13 P. aeruginosa and 11 K. pneumoniae phages of 18 different genera, whose host range was grouped as 1.2–17%, 28–48% and 60–87%, using a large collection of P. aeruginosa (n = 102) and K. pneumoniae (n = 155) strains carrying different virulence factors and phage binding receptors. We introduced the interpretation model curve for phage liquid culturing, which allows easy and quick analysis of bacterial and phage co-proliferation and growth of phage-resistant mutants (PRM) based on qualitative and partially quantitative evaluations. We assayed phage lytic activities both individually and in 14 different cocktails on planktonic bacterial cultures, including three resistotypes of P. aeruginosa (PAO1, PA14 and PA7) and seven K. pneumoniae strains of different capsular serotypes. Based on the results, the natural phage cocktails designed and tested in this study largely performed well and inhibited PRM growth either synergistically or in proto-cooperation. This study contributes to the knowledge of phage behavior in cocktails and the formulation of therapeutic phage preparations. The paper also provides a detailed description of the methods of working with phages.
Full article
(This article belongs to the Special Issue Bacteriophages and Other Alternative Antimicrobials to Combat Multidrug Resistance)
►▼
Show Figures
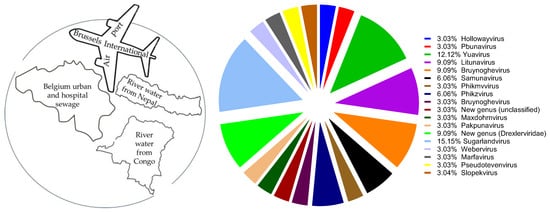
Figure 1
Open AccessEditorial
Bacterial and Fungal Pathogens: New Weapons to Fight Them
by
Charlotte A. Huber
Antibiotics 2024, 13(5), 384; https://doi.org/10.3390/antibiotics13050384 - 24 Apr 2024
Abstract
In high-income countries, degenerative diseases are the primary cause of death [...]
Full article
(This article belongs to the Special Issue Molecular Methods in Antibiotics Discovery)
Open AccessArticle
Association between Empirical Anti-Pseudomonal Antibiotics and Progression to Thoracic Surgery and Death in Empyema: Database Research
by
Akihiro Shiroshita, Kentaro Tochitani, Yohei Maki, Takero Terayama and Yuki Kataoka
Antibiotics 2024, 13(5), 383; https://doi.org/10.3390/antibiotics13050383 - 24 Apr 2024
Abstract
Evidence on the optimal antibiotic strategy for empyema is lacking. Our database study aimed to evaluate the effectiveness of empirical anti-pseudomonal antibiotics in patients with empyema. We utilised a Japanese real-world data database, focusing on patients aged ≥40 diagnosed with empyema, who underwent
[...] Read more.
Evidence on the optimal antibiotic strategy for empyema is lacking. Our database study aimed to evaluate the effectiveness of empirical anti-pseudomonal antibiotics in patients with empyema. We utilised a Japanese real-world data database, focusing on patients aged ≥40 diagnosed with empyema, who underwent thoracostomy and received intravenous antibiotics either upon admission or the following day. Patients administered intravenous vasopressors were excluded. We compared thoracic surgery and death within 90 days after admission between patients treated with empirical anti-pseudomonal and non-anti-pseudomonal antibiotics. Cause-specific hazard ratios for thoracic surgery and death were estimated using Cox proportional hazards models, with adjustment for clinically important confounders. Subgroup analyses entailed the same procedures for patients exhibiting at least one risk factor for multidrug-resistant organisms. Between March 2014 and March 2023, 855 patients with empyema meeting the inclusion criteria were enrolled. Among them, 271 (31.7%) patients received anti-pseudomonal antibiotics. The Cox proportional hazards models indicated that compared to empirical non-anti-pseudomonal antibiotics, empirical anti-pseudomonal antibiotics were associated with higher HRs for thoracic surgery and death within 90 days, respectively. Thus, regardless of the risks of multidrug-resistant organisms, empirical anti-pseudomonal antibiotics did not extend the time to thoracic surgery or death within 90 days.
Full article
(This article belongs to the Special Issue Antimicrobial Treatment of Lower Respiratory Tract Infections)
►▼
Show Figures
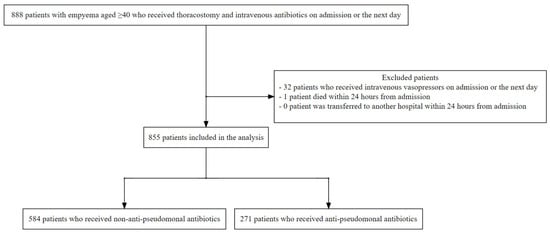
Figure 1
Open AccessArticle
Online-Assisted Survey on Antibiotic Use by Pet Owners in Dogs and Cats
by
Clara Rocholl, Yury Zablotski and Bianka Schulz
Antibiotics 2024, 13(5), 382; https://doi.org/10.3390/antibiotics13050382 - 24 Apr 2024
Abstract
The aim of the study was two-fold: first, to collect data on the use of antibiotics in Germany for dogs and cats and, second, their owners’ experiences and opinions. Using an anonymous online survey, dog and cat owners were asked about the last
[...] Read more.
The aim of the study was two-fold: first, to collect data on the use of antibiotics in Germany for dogs and cats and, second, their owners’ experiences and opinions. Using an anonymous online survey, dog and cat owners were asked about the last antibiotic administration in their pet. The inclusion criterion was any antibiotic administration within the last year. A total of 708 questionnaires from 463 dogs and 245 cats could be evaluated. Diarrhea was reported as the most common reason for antibiotic administration in dogs (18.4%). Wound infection/abscess/bite injury was the second most common reason in dogs (16.0%). In cats wound infection/abscess/bite injury was the most common reason (23.3%), followed by dental treatment (21.2%) and upper respiratory tract infections (16.7%). The most common antibiotics used systemically in both species were amoxicillin/clavulanic acid (32.5%), amoxicillin (14.8%), metronidazole (6.9%), and doxycycline (6.8%). While efficacy (99.9%) and tolerability (94.8%) were rated as most important for the choice of antibiotics, costs (51.6%) were cited as predominantly unimportant. First-line antibiotics were used significantly more often than critically important antibiotics. The majority of animal owners show awareness for avoidance of antibiotic resistance and the use of critically important antibiotics.
Full article
(This article belongs to the Section Antibiotics in Animal Health)
►▼
Show Figures
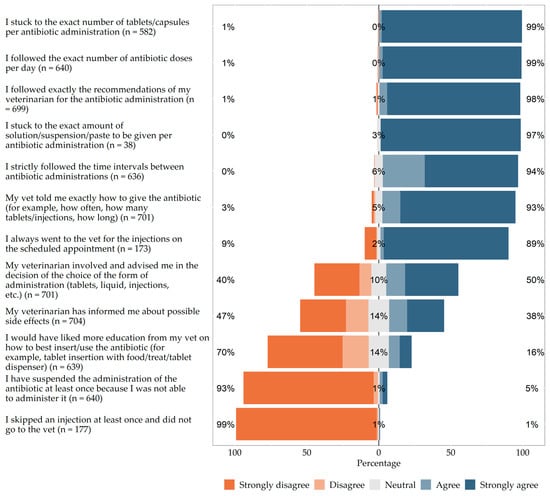
Figure 1
Open AccessArticle
Multidrug-Resistant Escherichia coli Remains Susceptible to Metal Ions and Graphene-Based Compounds
by
Nathalie Karaky, Shiying Tang, Parameshwari Ramalingam, Andrew Kirby, Andrew J. McBain, Craig E. Banks and Kathryn A. Whitehead
Antibiotics 2024, 13(5), 381; https://doi.org/10.3390/antibiotics13050381 - 24 Apr 2024
Abstract
Escherichia coli is listed as a priority 1 pathogen on the World Health Organization (WHO) priority pathogen list. For this list of pathogens, new antibiotics are urgently needed to control the emergence and spread of multidrug-resistant strains. This study assessed eighteen metal ions, graphene,
[...] Read more.
Escherichia coli is listed as a priority 1 pathogen on the World Health Organization (WHO) priority pathogen list. For this list of pathogens, new antibiotics are urgently needed to control the emergence and spread of multidrug-resistant strains. This study assessed eighteen metal ions, graphene, and graphene oxide for their antimicrobial efficacy against E. coli in both planktonic and biofilm growth states and the potential synergy between metal ions and graphene-based compounds. Molybdenum and tin ions exhibited the greatest antimicrobial activity against the planktonic states of the isolates with minimal inhibitory concentrations (MIC) ranging between 13 mg/L and 15.6 mg/L. Graphene oxide had no antimicrobial effect against any of the isolates, while graphene showed a moderate effect against E. coli (MIC, 62.5 mg/L). Combinations of metal ions and graphene-based compounds including tin–graphene, tin–graphene oxide, gold–graphene, platinum–graphene, and platinum–graphene oxide exhibited a synergistic antimicrobial effect (FIC ≤ 0.5), inhibiting the planktonic and biofilm formation of the isolates regardless of their antibiotic-resistant profiles. The bactericidal effect of the metal ions and the synergistic effects when combined with graphene/graphene oxide against medically relevant pathogens demonstrated that the antimicrobial efficacy was increased. Hence, such agents may potentially be used in the production of novel antimicrobial/antiseptic agents.
Full article
(This article belongs to the Special Issue Antibiotic Adjuvants: An Approach to Overcoming Multi-Drug Resistance and Biofilm Infections)
►▼
Show Figures
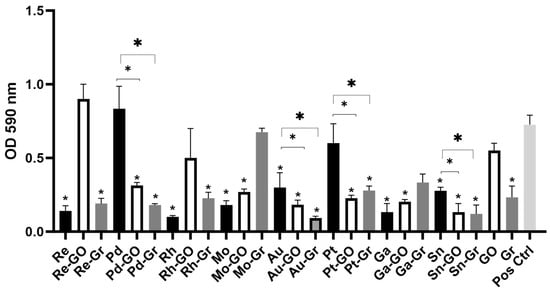
Figure 1
Open AccessArticle
Camel Milk Resistome in Kuwait: Genotypic and Phenotypic Characterization
by
Rita Rahmeh, Abrar Akbar, Batlah Almutairi, Mohamed Kishk, Naida Babic Jordamovic, Abdulaziz Al-Ateeqi, Anisha Shajan, Heba Al-Sherif, Alfonso Esposito, Sabah Al-Momin and Silvano Piazza
Antibiotics 2024, 13(5), 380; https://doi.org/10.3390/antibiotics13050380 - 23 Apr 2024
Abstract
Antimicrobial resistance (AMR) is one of the major global health and economic threats. There is growing concern about the emergence of AMR in food and the possibility of transmission of microorganisms possessing antibiotic resistance genes (ARGs) to the human gut microbiome. Shotgun sequencing
[...] Read more.
Antimicrobial resistance (AMR) is one of the major global health and economic threats. There is growing concern about the emergence of AMR in food and the possibility of transmission of microorganisms possessing antibiotic resistance genes (ARGs) to the human gut microbiome. Shotgun sequencing and in vitro antimicrobial susceptibility testing were used in this study to provide a detailed characterization of the antibiotic resistance profile of bacteria and their ARGs in dromedary camel milk. Eight pooled camel milk samples, representative of multiple camels distributed in the Kuwait desert, were collected from retail stores and analyzed. The genotypic analysis showed the presence of ARGs that mediate resistance to 18 classes of antibiotics in camel milk, with the highest resistance to fluoroquinolones (12.48%) and disinfecting agents and antiseptics (9%). Furthermore, the results pointed out the possible transmission of the ARGs to other bacteria through mobile genetic elements. The in vitro antimicrobial susceptibility testing indicated that 80% of the isolates were resistant to different classes of antibiotics, with the highest resistance observed against three antibiotic classes: penicillin, tetracyclines, and carbapenems. Multidrug-resistant pathogens including Klebsiella pneumoniae, Escherichia coli, and Enterobacter hormaechei were also revealed. These findings emphasize the human health risks related to the handling and consumption of raw camel milk and highlight the necessity of improving the hygienic practices of farms and retail stores to control the prevalence of ARGs and their transmission.
Full article
(This article belongs to the Special Issue Antimicrobial Resistance in Animals and Animal Products and Its Environmental Transmission Aspects)
►▼
Show Figures
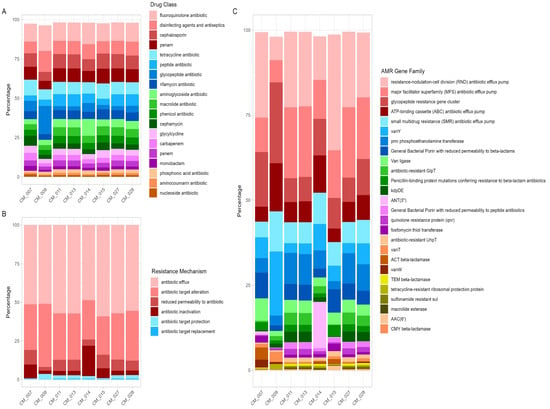
Figure 1

Journal Menu
► ▼ Journal Menu-
- Antibiotics Home
- Aims & Scope
- Editorial Board
- Reviewer Board
- Topical Advisory Panel
- Instructions for Authors
- Special Issues
- Topics
- Sections & Collections
- Article Processing Charge
- Indexing & Archiving
- Editor’s Choice Articles
- Most Cited & Viewed
- Journal Statistics
- Journal History
- Journal Awards
- Conferences
- Editorial Office
Journal Browser
► ▼ Journal BrowserHighly Accessed Articles
Latest Books
E-Mail Alert
News
Topics
Topic in
Antibiotics, Antioxidants, JoF, Microbiology Research, Microorganisms
Redox in Microorganisms, 2nd Edition
Topic Editors: Michal Letek, Volker BehrendsDeadline: 31 July 2024
Topic in
Antibiotics, JPM, Pharmaceuticals, Pharmaceutics
Pharmacokinetic and Pharmacodynamic Modelling in Drug Discovery and Development
Topic Editors: Inaki F. Troconiz, Victor Mangas Sanjuán, Maria Garcia-Cremades MiraDeadline: 31 August 2024
Topic in
Molecules, Pharmaceutics, Antibiotics, Microorganisms, Biomolecules, Marine Drugs, Polymers, IJMS
Antimicrobial Agents and Nanomaterials
Topic Editors: Sandra Pinto, Vasco D. B. BonifácioDeadline: 30 September 2024
Topic in
Antibiotics, Biomedicines, JCM, Pharmaceuticals, Pharmaceutics
Challenges and Future Prospects of Antibacterial Therapy
Topic Editors: Kwang-sun Kim, Zehra EdisDeadline: 31 October 2024

Conferences
Special Issues
Special Issue in
Antibiotics
When Communities Matter: Interplay between Mobile Genetic Elements and Antibiotic Resistance
Guest Editor: Antony T. VincentDeadline: 30 April 2024
Special Issue in
Antibiotics
Antibacterial Treatment in Periodontal and Endodontic Therapy, 2nd Volume
Guest Editors: Felix Krause, Andreas BraunDeadline: 15 May 2024
Special Issue in
Antibiotics
The Application of Antibiotic Therapy in Oral Surgery and Dental Implant Procedures
Guest Editors: Angel-Orion Salgado-Peralvo, Juan Francisco Peña-Cardelles, Eugenio Velasco-OrtegaDeadline: 31 May 2024
Special Issue in
Antibiotics
Challenges for Therapeutic Drug Monitoring of Antimicrobials
Guest Editors: Giuseppe Nunnari, Fabrizio Taglietti, Giuseppe PipitoneDeadline: 15 June 2024
Topical Collections
Topical Collection in
Antibiotics
Antimicrobial Resistance and Anti-Biofilms
Collection Editors: Ding-Qiang Chen, Yulong Tan, Ren-You Gan, Guanggang Qu, Zhenbo Xu, Junyan Liu