microRNAs in Health and Diseases
A topical collection in Cells (ISSN 2073-4409). This collection belongs to the section "Cell Signaling".
Viewed by 68537Editor
Topical Collection Information
Dear Colleagues,
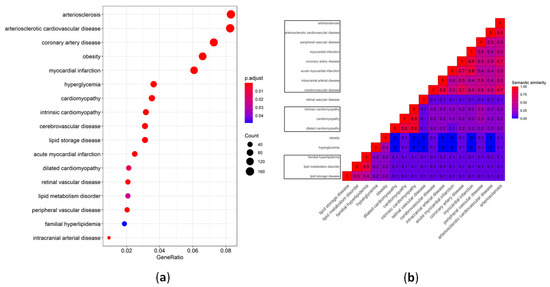
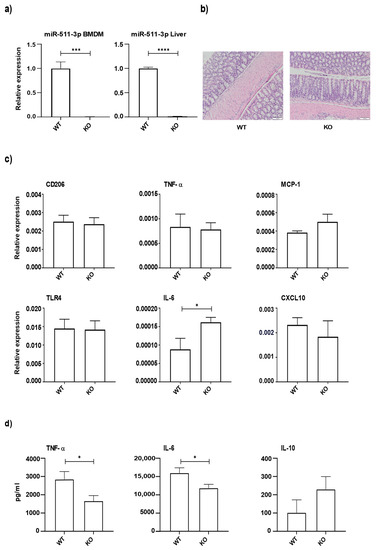
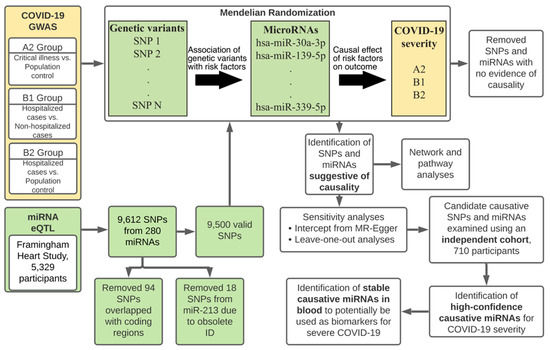
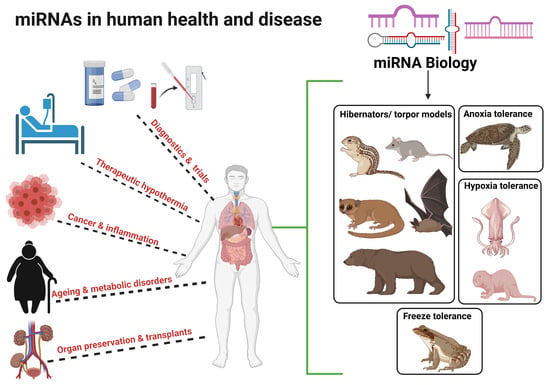
The microRNAs are small non-coding RNAs that regulate the expression of target genes by binding to the 3' untranslated region of the mRNAs. A single gene can be targeted by hundreds of miRs and a single miR can target hundreds of genes. This complex system makes it difficult to identify the biologically significant miR-mRNA interaction having a functional influence. Nonetheless, I am amazed to see the upcoming research that describes the miR-mRNA interactions in health and disease. The specific and efficient targetability of miRs makes them an attractive drug target. The miRs role in health and disease can be broadly divided into a) identification of biologically significant miR-gene interaction, b) development of efficient modulators with minimal non-specific binding, and c) testing the safety and toxicological characteristics of these molecules. The miRs that are expressed in a tissue-selective fashion (e.g., liver; miR-122, heart; miR-208, islets; miR-375), lifestyle modifications that affect the miR profile (e.g., diet, gut microbiota, fasting), novel molecules with better efficiency at miR-targeting and evaluation of their biocompatibility falls within the scope of this collection. I believe that an understanding of the extent to which miR(s) contribute to the development of the disease can source novel drug targets. In this collection, we aim to update the current understanding of the role of miRs in health and disease. I look forward to your contributions.
Dr. Ajit Vikram
Collection Editor
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Cells is an international peer-reviewed open access semimonthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript. The Article Processing Charge (APC) for publication in this open access journal is 2700 CHF (Swiss Francs). Submitted papers should be well formatted and use good English. Authors may use MDPI's English editing service prior to publication or during author revisions.
Keywords
- microRNAs
- Sirtuins
- tissue-specific
- miR-mRNA interaction
- heart failure
- hypertension
- nucleic-acids
- biocompatibility