Single- and Multiple-Adulterants Determinations of Goat Milk Powder by NIR Spectroscopy Combined with Chemometric Algorithms
Abstract
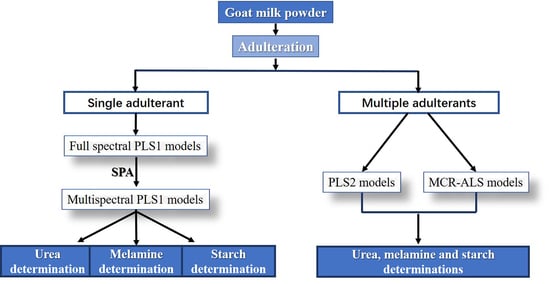
:1. Introduction
2. Materials and Methods
2.1. Sample Preparations
2.2. Acquisition of NIR Spectra Data
2.3. Pretreatment and Successive Projections Algorithm
2.4. Partial Least Squares Regression (PLS)
2.5. Multivariate Curve Resolution Alternating Least Squares (MCR-ALS)
3. Results and Discussion
3.1. Raw Spectral Characteristic Analysis
3.2. Adulteration Analysis of Single Adulterant
3.2.1. Spectral Pretreatment and Full Spectral Model
3.2.2. Multispectral Models Optimized by SPA
3.3. Adulteration Analysis of Multiple Adulterants
3.3.1. PLS2 Models
3.3.2. MCR-ALS Models
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hodgkinson, A.J.; Wallace, O.A.; Boggs, I.; Broadhurst, M.; Prosser, C.G. Gastric digestion of cow and goat milk: Impact of infant and young child in vitro digestion conditions. Food Chem. 2018, 245, 275–281. [Google Scholar] [CrossRef] [PubMed]
- Verruck, S.; Dantas, A.; Prudencio, E.S. Functionality of the components from goat’s milk, recent advances for functional dairy products development and its implications on human health. J. Funct. Foods 2019, 52, 243–257. [Google Scholar] [CrossRef]
- Nuñez, M.; de Renobales, M. IDF international symposium on sheep, goat and other non-cow milk. Int. Dairy J. 2016, 100, 1. [Google Scholar] [CrossRef]
- Ranadheera, C.S.; Evans, C.A.; Baines, S.K.; Balthazar, C.F.; Cruz, A.G.; Esmerino, E.A.; Freitas, M.Q.; Pimentel, T.C.; Wittwer, A.E.; Naumovski, N.; et al. Probiotics in goat milk products: Delivery capacity and ability to improve sensory attributes. Compr. Rev. Food Sci. Food Saf. 2019, 18, 867–882. [Google Scholar] [CrossRef] [Green Version]
- Pulina, G.; Milán, M.J.; Lavín, M.P.; Theodoridis, A.; Morin, E.; Capote, J.; Thomas, D.L.; Francesconi, A.H.D.; Caja, G. Invited review: Current production trends, farm structures, and economics of the dairy sheep and goat sectors. J. Dairy Sci. 2018, 101, 6715–6729. [Google Scholar] [CrossRef] [Green Version]
- Cattaneo, T.M.; Holroyd, S.E. The use of near infrared spectroscopy for determination of adulteration and contamination in milk and milk powder: Updating knowledge. J. Near Infrared Spectrosc. 2013, 21, 341–349. [Google Scholar] [CrossRef]
- Da Paixao Teixeira, J.L.; dos Santos Carames, E.T.; Baptista, D.P.; Gigante, M.L.; Pallone, J.A.L. Vibrational spectroscopy and chemometrics tools for authenticity and improvement the safety control in goat milk. Food Control 2020, 112, 107105. [Google Scholar] [CrossRef]
- Ke, X.; Zhang, J.; Lai, S.; Chen, Q.; Zhang, Y.; Jiang, Y.; Mo, W.; Ren, Y. Quantitative analysis of cow whole milk and whey powder adulteration percentage in goat and sheep milk products by isotopic dilution-ultra-high performance liquid chromatography-tandem mass spectrometry. Anal. Bioanal. Chem. 2017, 409, 213–224. [Google Scholar] [CrossRef]
- Núñez, O. The Role of Capillary Electrophoresis in Food Integrity and Authenticity. In Chromatographic and Related Separation Techniques in Food Integrity and Authenticity. Volume A: Advances in Chromatographic Techniques; World Scientific Publishing Co Pte Ltd.: Singapore, 2021; pp. 71–104. [Google Scholar]
- Galan-Malo, P.; Mendiara, I.; Razquin, P.; Mata, L. Validation of a rapid lateral flow method for the detection of cows’ milk in water buffalo, sheep or goat milk. Food Addit. Contam. Part A 2018, 35, 609–614. [Google Scholar] [CrossRef]
- Di Pinto, A.; Terio, V.; Marchetti, P.; Bottaro, M.; Mottola, A.; Bozzo, G.; Bonerba, E.; Ceci, E.; Tantillo, G. DNA-based approach for species identification of goat-milk products. Food Chem. 2017, 229, 93–97. [Google Scholar] [CrossRef]
- De Brito, A.A.; Campos, F.; dos Reis Nascimento, A.; Damiani, C.; da Silva, F.A.; de Almeida Teixeira, G.H.; Júnior, L.C.C. Non-destructive determination of color, titratable acidity, and dry matter in intact tomatoes using a portable Vis-NIR spectrometer. J. Food Compos. Anal. 2021, 107, 104288. [Google Scholar] [CrossRef]
- Wang, J.; Zareef, M.; He, P.; Sun, H.; Chen, Q.; Li, H.; Ouyang, Q.; Guo, Z.; Zhang, Z.; Xu, D. Evaluation of matcha tea quality index using portable NIR spectroscopy coupled with chemometric algorithms. J. Sci. Food Agric. 2019, 99, 5019–5027. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.W.; Hwang, U.T.; Jo, K.; Lee, T.; Park, J.; Kim, J.C.; Kwak, H.W.; Choi, I.; Yeo, H. NIR-chemometric approaches for evaluating carbonization characteristics of hydrothermally carbonized lignin. Sci. Rep. 2021, 11, 1–8. [Google Scholar] [CrossRef]
- Deidda, R.; Sacre, P.Y.; Clavaud, M.; Coïc, L.; Avohou, H.; Hubert, P.; Ziemons, E. Vibrational spectroscopy in analysis of pharmaceuticals: Critical review of innovative portable and handheld NIR and Raman spectrophotometers. TrAC Trends Anal. Chem. 2019, 114, 251–259. [Google Scholar] [CrossRef]
- Li, X.; Zhang, L.; Zhang, Y.; Wang, D.; Wang, X.; Yu, L.; Zhang, W.; Li, P. Review of NIR spectroscopy methods for nondestructive quality analysis of oilseeds and edible oils. Trends Food Sci. Technol. 2020, 101, 172–181. [Google Scholar] [CrossRef]
- Karunathilaka, S.R.; Yakes, B.J.; He, K.; Chung, J.K.; Mossoba, M. Non-targeted NIR spectroscopy and SIMCA classification for commercial milk powder authentication: A study using eleven potential adulterants. Heliyon 2018, 4, e00806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Da Silva Dias, L.; da Silva Junior, J.C.; Felício, A.L.D.S.M.; de França, J.A. A NIR photometer prototype with integrating sphere for the detection of added water in raw milk. IEEE Trans. Instrum. Meas. 2018, 67, 2812–2819. [Google Scholar] [CrossRef]
- Windarsih, A.; Rohman, A.; Irnawati; Riyanto, S. The Combination of Vibrational Spectroscopy and Chemometrics for Analysis of Milk Products Adulteration. Int. J. Food Sci. 2021, 2021, 8853358. [Google Scholar] [CrossRef]
- Da Paixao Teixeira, J.L.; dos Santos Carames, E.T.; Baptista, D.P.; Gigante, M.L.; Pallone, J.A.L. Rapid adulteration detection of yogurt and cheese made from goat milk by vibrational spectroscopy and chemometric tools. J. Food Compos. Anal. 2021, 96, 103712. [Google Scholar] [CrossRef]
- Dos Santos Pereira, E.V.; de Sousa Fernandes, D.D.; de Araújo, M.C.U.; Diniz, P.H.G.D.; Maciel, M.I.S. Simultaneous determination of goat milk adulteration with cow milk and their fat and protein contents using NIR spectroscopy and PLS algorithms. LWT 2020, 127, 109427. [Google Scholar] [CrossRef]
- Teixeira, J.L.D.P.; Caramês, E.T.D.S.; Baptista, D.P.; Gigante, M.L.; Pallone, J.A.L. Adulteration Detection in Goat Dairy Beverage Through NIR Spectroscopy and DD-SIMCA. Food Anal. Methods 2021, 15, 783–791. [Google Scholar] [CrossRef]
- Barnes, R.J.; Dhanoa, M.S.; Lister, S.J. Standard normal variate transformation and de-trending of near-infrared diffuse reflectance spectra. Appl. Spectrosc. 1989, 43, 772–777. [Google Scholar] [CrossRef]
- Lu, B.; Wang, X.; Liu, N.; He, K.; Wu, K.; Li, H.; Tang, X. Feasibility of NIR spectroscopy detection of moisture content in coco-peat substrate based on the optimization characteristic variables. Spectrochim. Acta Part A 2020, 239, 118455. [Google Scholar] [CrossRef] [PubMed]
- Stocchero, M. Iterative deflation algorithm, eigenvalue equations, and PLS2. J. Chemom. 2019, 33, e3144. [Google Scholar] [CrossRef]
- Galtier, O.; Abbas, O.; Le Dréau, Y.; Rebufa, C.; Kister, J.; Artaud, J.; Dupuy, N. Comparison of PLS1-DA, PLS2-DA and SIMCA for classification by origin of crude petroleum oils by MIR and virgin olive oils by NIR for different spectral regions. Vib. Spectrosc. 2011, 55, 132–140. [Google Scholar] [CrossRef]
- Pedro, A.M.; Ferreira, M.M. Simultaneously calibrating solids, sugars and acidity of tomato products using PLS2 and NIR spectroscopy. Anal. Chim. Acta 2007, 595, 221–227. [Google Scholar] [CrossRef]
- De Oliveira Neves, A.C.; Zougagh, M.; Ríos, Á.; Tauler, R.; Wakamatsu, K.; Galván, I. Pheomelanin subunit non-destructive quantification by Raman spectroscopy and multivariate curve resolution-alternating least squares (MCR-ALS). Chemom. Intell. Lab. Syst. 2021, 217, 104406. [Google Scholar] [CrossRef]
- Stella, A.; Bonnier, F.; Tfayli, A.; Yvergnaux, F.; Byrne, H.J.; Chourpa, I.; Munnier, E.; Tauber, C. Raman mapping coupled to self-modelling MCR-ALS analysis to estimate active cosmetic ingredient penetration profile in skin. J. Biophotonics 2020, 13, e202000136. [Google Scholar] [CrossRef]
- Mazivila, S.J.; Lombardi, J.M.; Páscoa, R.N.; Bortolato, S.A.; Leitão, J.M.; da Silva, J.C.E. Three-way calibration using PARAFAC and MCR-ALS with previous synchronization of second-order chromatographic data through a new functional alignment of pure vectors for the quantification in the presence of retention time shifts in peak position and shape. Anal. Chim. Acta 2021, 1146, 98–108. [Google Scholar] [CrossRef]
- Alcaráz, M.R.; Schenone, A.V.; Culzoni, M.J.; Goicoechea, H.C. Modeling of second-order spectrophotometric data generated by a pH-gradient flow injection technique for the determination of doxorubicin in human plasma. Microchem. J. 2014, 112, 25–33. [Google Scholar] [CrossRef]
- Kaur, P.; Imteaz, M.A.; Sillanpää, M.; Sangal, V.K.; Kushwaha, J.P. Parametric optimization and MCR-ALS kinetic modeling of electro oxidation process for the treatment of textile wastewater. Chemom. Intell. Lab. Syst. 2020, 203, 104027. [Google Scholar] [CrossRef]
- De Meutter, J.; Goormaghtigh, E. Protein Structural Denaturation Evaluated by MCR-ALS of Protein Microarray FTIR Spectra. Anal. Chem. 2021, 93, 13441–13449. [Google Scholar] [CrossRef] [PubMed]
- Benavente, F.; Pero-Gascon, R.; Pont, L.; Jaumot, J.; Barbosa, J.; Sanz-Nebot, V. Identification of antihypertensive peptides in nutraceuticals by capillary electrophoresis-mass spectrometry. J. Chromatogr. A 2018, 1579, 129–137. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, G.; Zhou, X.; Li, Y. Determination of acetamiprid partial-intercalative binding to DNA by use of spectroscopic, chemometrics, and molecular docking techniques. Anal. Bioanal. Chem. 2013, 405, 8871–8883. [Google Scholar] [CrossRef]
- Azzouz, T.; Tauler, R. Application of multivariate curve resolution alternating least squares (MCR-ALS) to the quantitative analysis of pharmaceutical and agricultural samples. Talanta 2008, 74, 1201–1210. [Google Scholar] [CrossRef] [PubMed]
- Castro, R.C.; Ribeiro, D.S.; Santos, J.L.; Páscoa, R.N. Near infrared spectroscopy coupled to MCR-ALS for the identification and quantification of saffron adulterants: Application to complex mixtures. Food Control 2021, 123, 107776. [Google Scholar] [CrossRef]
- Colares, C.J.; Pastore, T.C.; Coradin, V.T.; Marques, L.F.; Moreira, A.C.; Alexandrino, G.L.; Poppi, R.J.; Braga, J.W. Near infrared hyperspectral imaging and MCR-ALS applied for mapping chemical composition of the wood specie Swietenia Macrophylla King (Mahogany) at microscopic level. Microchem. J. 2016, 124, 356–363. [Google Scholar] [CrossRef]
- González-Sáiz, J.M.; Esteban-Díez, I.; Rodríguez-Tecedor, S.; Pizarro, C. Valorization of onion waste and by-products: MCR-ALS applied to reveal the compositional profiles of alcoholic fermentations of onion juice monitored by near-infrared spectroscopy. Biotechnol. Bioeng. 2008, 101, 776–787. [Google Scholar] [CrossRef]
- Jaumot, J.; de Juan, A.; Tauler, R. MCR-ALS GUI 2.0: New features and applications. Chemom. Intell. Lab. Syst. 2015, 140, 1–12. [Google Scholar] [CrossRef]
- Dos Santos Pereira, E.V.; de Sousa Fernandes, D.D.; de Araújo, M.C.U.; Diniz, P.H.G.D.; Maciel, M.I.S. In-situ authentication of goat milk in terms of its adulteration with cow milk using a low-cost portable NIR spectrophotometer. Microchem. J. 2021, 163, 105885. [Google Scholar] [CrossRef]
- Yun, Y.H.; Li, H.D.; Deng, B.C.; Cao, D.S. An overview of variable selection methods in multivariate analysis of near-infrared spectra. TrAC Trends Anal. Chem. 2019, 113, 102–115. [Google Scholar] [CrossRef]
- Masemola, C.; Cho, M.A. Estimating leaf nitrogen concentration from similarities in fresh and dry leaf spectral bands using a model population analysis framework. Int. J. Remote Sens. 2019, 40, 6841–6860. [Google Scholar] [CrossRef]
- Qiu, G.; Lü, E.; Wang, N.; Lu, H.; Wang, F.; Zeng, F. Cultivar classification of single sweet corn seed using fourier transform near-infrared spectroscopy combined with discriminant analysis. Appl. Sci. 2019, 9, 1530. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.C.; Yoon, Y.H.; Kim, S.M.; Pyo, B.S.; Hsieh, F.H.; Kim, H.J.; Eun, J.B. Rapid prediction of amylose content of polished rice by Fourier transform near-infrared spectroscopy. Food Sci. Biotechnol. 2007, 16, 477–481. [Google Scholar]










| Number | Goat Milk Powder | Urea | Melamine | Starch |
|---|---|---|---|---|
| 1 | 98.5% | 0% | 0.5% | 1% |
| 2 | 97.7% | 0.8% | 1.5% | 0% |
| 3 | 97.3% | 1.5% | 0% | 1.2% |
| 4 | 92% | 1% | 2% | 5% |
| 5 | 84% | 2% | 4% | 10% |
| 6 | 72% | 5% | 8% | 15% |
| 7 | 60% | 8% | 12% | 20% |
| Actual Value (%) | Predicted Value (%) | Absolute error (%) | ||||||
|---|---|---|---|---|---|---|---|---|
| Urea | Melamine | Starch | Urea | Melamine | Starch | Urea | Melamine | Starch |
| 0.0 | 0.00 | 0.0 | 0.0 | −0.01 | −0.1 | 0.0 | 0.01 | 0.1 |
| 0.5 | 0.01 | 1.0 | 0.3 | −0.00 | 1.1 | 0.2 | 0.01 | 0.1 |
| 0.8 | 0.05 | 5.0 | 0.9 | 0.04 | 4.9 | 0.1 | 0.01 | 0.1 |
| 1.0 | 0.10 | 10.0 | 1.4 | 0.13 | 9.9 | 0.4 | 0.03 | 0.1 |
| 2.0 | 0.50 | 15.0 | 2.2 | 0.47 | 15.2 | 0.2 | 0.03 | 0.2 |
| 5.0 | 1.00 | 20.0 | 5.3 | 1.02 | 20.1 | 0.3 | 0.02 | 0.1 |
| 8.0 | 5.00 | 25.0 | 7.7 | 5.05 | 25.0 | 0.3 | 0.05 | 0.0 |
| 10.0 | 10.00 | 30.0 | 10.0 | 9.98 | 29.9 | 0.0 | 0.02 | 0.1 |
| Adulterant | Wavelength (nm) |
|---|---|
| urea | 993, 1037, 1219 |
| melamine | 1027, 1929, 2208 |
| starch | 802, 1612, 1724, 1873 |
| Adulterant | LVs | Calibration | Cross-Validation | Prediction | |||
|---|---|---|---|---|---|---|---|
| RMSEC (%) | RMSECV (%) | RMSEP (%) | |||||
| urea | 3 | 0.990 | 0.361 | 0.989 | 0.369 | 0.987 | 0.403 |
| melamine | 3 | 0.999 | 0.129 | 0.999 | 0.129 | 0.999 | 0.133 |
| starch | 4 | 1.000 | 0.210 | 1.000 | 0.213 | 1.000 | 0.220 |
| Pretreatment | Component | LVs | Calibration | Cross-Validation | Prediction | |||
|---|---|---|---|---|---|---|---|---|
| RMSEC (%) | RMSECV (%) | RMSEP (%) | ||||||
| NON | goat milk | 6 | 0.999 | 0.489 | 0.999 | 0.499 | 0.999 | 0.541 |
| urea | 6 | 0.999 | 0.099 | 0.998 | 0.106 | 0.998 | 0.110 | |
| melamine | 6 | 0.998 | 0.180 | 0.998 | 0.182 | 0.998 | 0.188 | |
| starch | 6 | 0.999 | 0.271 | 0.999 | 0.275 | 0.998 | 0.289 | |
| NOR | goat milk | 6 | 0.999 | 0.488 | 0.999 | 0.499 | 0.999 | 0.538 |
| urea | 6 | 0.999 | 0.096 | 0.999 | 0.102 | 0.998 | 0.111 | |
| melamine | 6 | 0.998 | 0.178 | 0.998 | 0.181 | 0.998 | 0.189 | |
| starch | 6 | 0.999 | 0.268 | 0.999 | 0.273 | 0.998 | 0.292 | |
| MAS | goat milk | 6 | 0.999 | 0.490 | 0.999 | 0.504 | 0.999 | 0.537 |
| urea | 6 | 0.998 | 0.111 | 0.998 | 0.115 | 0.998 | 0.126 | |
| melamine | 6 | 0.998 | 0.181 | 0.998 | 0.184 | 0.998 | 0.189 | |
| starch | 6 | 0.999 | 0.267 | 0.999 | 0.274 | 0.998 | 0.293 | |
| Pretreatment | Component | Number of Calibration/Prediction | Calibration | Prediction | |||||
|---|---|---|---|---|---|---|---|---|---|
| RMSEC (%) | RMSEP (%) | ||||||||
| NON | goat milk | 98/42 | 0.768 | 99.99 | 0.002 | 0.996 | 0.838 | 0.996 | 0.879 |
| urea | 0/140 | / | 0.679 | 2.529 | |||||
| melamine | 98/42 | 0.990 | 0.404 | 0.989 | 0.434 | ||||
| starch | 0/140 | / | 0.870 | 4.279 | |||||
| NOR | goat milk | 98/42 | 0.774 | 99.99 | 0.002 | 0.996 | 0.834 | 0.996 | 0.903 |
| urea | 0/140 | / | 0.681 | 2.368 | |||||
| melamine | 98/42 | 0.990 | 0.410 | 0.989 | 0.436 | ||||
| starch | 0/140 | / | 0.867 | 4.524 | |||||
| MAS | goat milk | 98/42 | 0.771 | 99.99 | 0.002 | 0.996 | 0.835 | 0.996 | 0.873 |
| urea | 0/140 | / | 0.686 | 2.604 | |||||
| melamine | 98/42 | 0.991 | 0.383 | 0.987 | 0.470 | ||||
| starch | 0/140 | / | 0.873 | 4.254 | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, X.; Wang, Y.; Liu, X.; Jiang, H.; Zhao, Z.; Niu, X.; Li, C.; Pang, B.; Li, Y. Single- and Multiple-Adulterants Determinations of Goat Milk Powder by NIR Spectroscopy Combined with Chemometric Algorithms. Agriculture 2022, 12, 434. https://doi.org/10.3390/agriculture12030434
Zhao X, Wang Y, Liu X, Jiang H, Zhao Z, Niu X, Li C, Pang B, Li Y. Single- and Multiple-Adulterants Determinations of Goat Milk Powder by NIR Spectroscopy Combined with Chemometric Algorithms. Agriculture. 2022; 12(3):434. https://doi.org/10.3390/agriculture12030434
Chicago/Turabian StyleZhao, Xin, Yunpeng Wang, Xin Liu, Hongzhe Jiang, Zhilei Zhao, Xiaoying Niu, Chunhua Li, Bin Pang, and Yanlei Li. 2022. "Single- and Multiple-Adulterants Determinations of Goat Milk Powder by NIR Spectroscopy Combined with Chemometric Algorithms" Agriculture 12, no. 3: 434. https://doi.org/10.3390/agriculture12030434