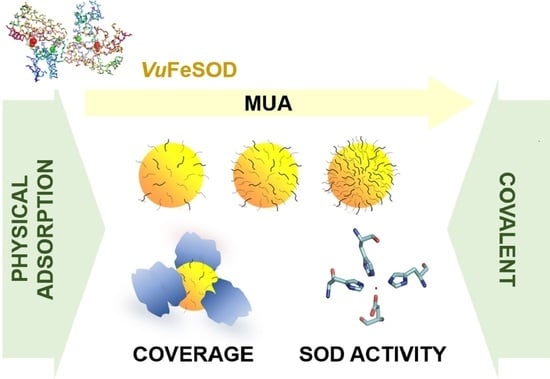
A Study of the Interface of Gold Nanoparticles Conjugated to Cowpea Fe-Superoxide Dismutase
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Biological Material
2.2. Methods
2.2.1. Overexpression, Purification and Activity of Recombinant VuFeSOD
2.2.2. Gold Nanoparticles Synthesis (AuNPs)
2.2.3. Surface Modification of AuNPs
2.2.4. Conjugation of SOD to AuNPs
2.2.5. Superoxide Dismutase In-Gel Activity Assays for the Conjugates
2.2.6. Spectrophotometric Activity Assay
2.2.7. Dynamic Light Scattering
3. Results and Discussion
3.1. Engineering the NP Surface Chemistry
3.2. Gold Nanoparticle Protein Conjugation
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Abreu, I.A.; Cabelli, D.E. Superoxide dismutases—A review of the metal-associated mechanistic variations. Biochim. Biophys. Acta Proteins Proteom. 2010, 1804, 263–274. [Google Scholar] [CrossRef] [PubMed]
- Halliwell, B.; Gutteridge, J.M.C. Free Radicals in Biology and Medicine. Free Radic. Biol. Med. 2015, 5, 961. [Google Scholar] [CrossRef]
- Gopalakrishnan, B.; Nash, K.M.; Velayutham, M.; Villamena, F.A. Detection of nitric oxide and superoxide radical anion by electron paramagnetic resonance spectroscopy from cells using spin traps. J. Vis. Exp. 2012, 66, e2810. [Google Scholar] [CrossRef] [Green Version]
- Jie, Z.; Liu, J.; Shu, M.; Ying, Y.; Yang, H. Detection strategies for superoxide anion: A review. Talanta 2022, 236, 122892. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Niu, X.; Zhao, H.; Tang, J.; Lan, M. Immobilization of superoxide dismutase on Pt-Pd/MWCNTs hybrid modified electrode surface for superoxide anion detection. Biosens. Bioelectron. 2015, 67, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Zhu, A.; Sun, K.; Petty, H.R. Titanium doping reduces superoxide dismutase activity, but not oxidase activity, of catalytic CeO2 nanoparticles. Inorg. Chem. Commun. 2012, 15, 235–237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, Z.; Tian, Y.; Yin, X.; Rui, Q.; Liu, H.; Luo, Y. Physical vapor deposited zinc oxide nanoparticles for direct electron transfer of superoxide dismutase. Electrochem. Commun. 2008, 10, 818–820. [Google Scholar] [CrossRef]
- Wang, L.; Mao, W.; Ni, D.; Di, J.; Wu, Y.; Tu, Y. Direct electrodeposition of gold nanoparticles onto indium/tin oxide film coated glass and its application for electrochemical biosensor. Electrochem. Commun. 2008, 10, 673–676. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, Y.; Wang, J.; Di, J. Disposable superoxide anion biosensor based on superoxide dismutase entrapped in silica sol-gel matrix at gold nanoparticles modified ITO electrode. Bioprocess Biosyst. Eng. 2009, 32, 531–536. [Google Scholar] [CrossRef]
- Wang, L.; Wen, W.; Xiong, H.; Zhang, X.; Gu, H.; Wang, S. A novel amperometric biosensor for superoxide anion based on superoxide dismutase immobilized on gold nanoparticle-chitosan-ionic liquid biocomposite film. Anal. Chim. Acta 2013, 758, 66–71. [Google Scholar] [CrossRef]
- El-Deab, M.S.; Ohsaka, T. Direct electron transfer of copper-zinc superoxide dismutase (SOD) on crystallographically oriented Au nanoparticles. Electrochem. Commun. 2007, 9, 651–656. [Google Scholar] [CrossRef]
- Hong, S.; Choi, I.; Lee, S.; Yang, Y.I.; Kang, T.; Yi, J. Sensitive and colorimetric detection of the structural evolution of superoxide dismutase with gold nanoparticles. Anal. Chem. 2009, 81, 1378–1382. [Google Scholar] [CrossRef] [PubMed]
- Kang, T.; Hong, S.; Choi, I.; Sung, J.J.; Kim, Y.; Hahn, J.S.; Yi, J. Reversible pH-driven conformational switching of tethered superoxide dismutase with gold nanoparticle enhanced surface plasmon resonance spectroscopy. J. Am. Chem. Soc. 2006, 128, 12870–12878. [Google Scholar] [CrossRef] [PubMed]
- Pudlarz, A.M.; Ranoszek-Soliwoda, K.; Czechowska, E.; Tomaszewska, E.; Celichowski, G.; Grobelny, J.; Szemraj, J. A Study of the Activity of Recombinant Mn-Superoxide Dismutase in the Presence of Gold and Silver Nanoparticles. Appl. Biochem. Biotechnol. 2019, 187, 1551–1568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tellechea, E.; Cornago, I.; Ciaurriz, P.; Moran, J.F.; Asensio, A.C. Conjugation of active iron superoxide dismutase to nanopatterned surfaces. IEEE Trans. Nanobioscience 2012, 11, 176–180. [Google Scholar] [CrossRef] [Green Version]
- Ge, B.; Scheller, F.W.; Lisdat, F. Electrochemistry of immobilized CuZnSOD and FeSOD and their interaction with superoxide radicals. Biosens. Bioelectron. 2002, 18, 295–302. [Google Scholar] [CrossRef]
- Asuri, P.; Karajanagi, S.S.; Vertegel, A.A.; Dordick, J.S.; Kane, R.S. Enhanced stability of enzymes adsorbed onto nanoparticles. J. Nanosci. Nanotechnol. 2007, 7, 1675–1678. [Google Scholar] [CrossRef]
- Lv, M.; Zhu, E.; Su, Y.; Li, Q.; Li, W.; Zhao, Y.; Huang, Q. Trypsin-gold nanoparticle conjugates: Binding, enzymatic activity, and stability. Prep. Biochem. Biotechnol. 2009, 39, 429–438. [Google Scholar] [CrossRef]
- Moran, J.F.; James, E.K.; Rubio, M.C.; Sarath, G.; Klucas, R.V.; Becana, M. Functional Characterization and Expression of a Cytosolic Iron-Superoxide Dismutase from Cowpea Root Nodules. Plant Physiol. 2003, 133, 773–782. [Google Scholar] [CrossRef] [Green Version]
- Asensio, A.C.; Gil-Monreal, M.; Pires, L.; Gogorcena, Y.; Aparicio-Tejo, P.M.; Moran, J.F. Two Fe-superoxide dismutase families respond differently to stress and senescence in legumes. J. Plant Physiol. 2012, 169, 1253–1260. [Google Scholar] [CrossRef]
- Muñoz, I.G.; Moran, J.F.; Becana, M.; Montoya, G. Crystallization and preliminary X-ray diffraction studies of the eukaryotic iron superoxide dismutase (FeSOD) from Vigna unguiculata. Acta Cryst. 2003, D59, 1070–1072. [Google Scholar] [CrossRef]
- Muñoz, I.G.; Moran, J.F.; Becana, M.; Montoya, G. The crystal structure of an eukaryotic iron superoxide dismutase suggests intersubunit cooperation during catalysis. Protein Sci. 2005, 14, 387–394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urarte, E.; Auzmendi, I.; Rol, S.; Ariz, I.; Aparicio-Tejo, P.; Arredondo-Peter, R.; Moran, J.F. A Self-Induction Method to Produce High Quantities of Recombinant Functional Flavo-Leghemoglobin Reductase. Methods Enzymol. 2008, 436, 411–423. [Google Scholar] [CrossRef]
- Ischiropoulos, H.; Zhu, L.; Chen, J.; Tsai, M.; Martin, J.C.; Smith, C.D.; Beckman, J.S. Peroxynitrite-mediated tyrosine nitration catalyzed by superoxide dismutase. Arch. Biochem. Biophys. 1992, 298, 431–437. [Google Scholar] [CrossRef]
- Ischiropoulos, H.; Zhu, L.; Beckman, J.S. Peroxynitrite formation from macrophage-derived nitric oxide. Arch. Biochem. Biophys. 1992, 298, 446–451. [Google Scholar] [CrossRef]
- Larrainzar, E.; Urarte, E.; Auzmendi, I.; Ariz, I.; Arrese-Igor, C.; González, E.M.; Moran, J.F. Use of Recombinant Iron-Superoxide Dismutase as A Marker of Nitrative Stress. Methods Enzymol. 2008, 437, 605–618. [Google Scholar] [PubMed]
- Urarte, E.; Asensio, A.C.; Tellechea, E.; Pires, L.; Moran, J.F. Evaluation of the anti-nitrative effect of plant antioxidants using a cowpea Fe-superoxide dismutase as a target. Plant Physiol. Biochem. 2014, 83, 356–364. [Google Scholar] [CrossRef]
- Beauchamp, C.; Fridovich, I. Superoxide dismutase: Improved assays and an assay applicable to acrylamide gels. Anal. Biochem. 1971, 44, 276–287. [Google Scholar] [CrossRef]
- Beesley, J.E. Colloidal gold: A new perspective for cytochemical marking. Trends Biochem. Sci. 1989, 14, 392. [Google Scholar] [CrossRef]
- Abràmoff, M.D.; Magalhães, P.J.; Ram, S.J. Image processing with ImageJ Part II. Biophotonics Int. 2005, 11, 36–43. [Google Scholar]
- Park, S.; Brown, K.A.; Hamad-Schifferli, K. Changes in oligonucleotide conformation on nanoparticle surfaces by modification with mercaptohexanol. Nano Lett. 2004, 4, 1925–1929. [Google Scholar] [CrossRef]
- Brown, K.A. Noncovalent Adsorption of Nucleotides in Gold Nanoparticle DNA Conjugates: Bioavailability at the Bio-Nano Interface. 2008. Available online: https://dspace.mit.edu/handle/1721.1/44866 (accessed on 11 October 2022).
- Sun, Y.; Oberley, L.W.; Li, Y. A simple method for clinical assay of superoxide dismutase. Clin. Chem. 1988, 34, 497–500. [Google Scholar] [CrossRef] [PubMed]
- Mie, G. Contributions to the optics of turbid media, particularly of colloidal metal solutions. Ann. Phys. 1976, 25, 377–445. [Google Scholar]
- Haynes, C.L.; van Duyne, R.P. Nanosphere lithography: A versatile nanofabrication tool for studies of size-dependent nanoparticle optics. J. Phys. Chem. B 2001, 105, 5599–5611. [Google Scholar] [CrossRef]
- Ivanov, M.R.; Haes, A.J. Anionic functionalized gold nanoparticle continuous full filling separations: Importance of sample concentration. Anal. Chem. 2012, 84, 1320–1326. [Google Scholar] [CrossRef]
- Hinterwirth, H.; Kappel, S.; Waitz, T.; Prohaska, T.; Lindner, W.; Lämmerhofer, M. Quantifying thiol ligand density of self-assembled monolayers on gold nanoparticles by inductively coupled plasma-mass spectrometry. ACS Nano 2013, 7, 1129–1136. [Google Scholar] [CrossRef]
- Reed, A.M.W.; Metallo, S.J. Oriented protein adsorption to gold nanoparticles through a genetically encodable binding motif. Langmuir 2010, 26, 18945–18950. [Google Scholar] [CrossRef]
- Sperling, R.A.; Parak, W.J. Surface modification, functionalization and bioconjugation of colloidal inorganic nanoparticles. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2010, 368, 1333–1383. [Google Scholar] [CrossRef]
- Pissuwan, D.; Cortie, C.H.; Valenzuela, S.M.; Cortie, M.B. Gold nanosphere-antibody conjugates for hyperthermal therapeutic applications. Gold Bull. 2007, 40, 121–129. [Google Scholar] [CrossRef] [Green Version]
- Hermanson, G.T. Chapter 4. Zero-Length Crosslinkers. In Bioconjugate Techniques, 3rd ed.; Academic Press: Cambridge, MA, USA, 2013; pp. 259–273. ISBN 9780123822390. [Google Scholar] [CrossRef]
- Getzoff, E.D.; Tainer, J.A.; Weiner, P.K.; Kollman, P.A.; Richardson, J.S.; Richardson, D.C. Electrostatic recognition between superoxide and copper, zinc superoxide dismutase. Nature 1983, 306, 287–290. [Google Scholar] [CrossRef]
- Aubin-Tam, M.E.; Hamad-Schifferli, K. Gold nanoparticle-cytochrome C complexes: The effect of nanoparticle ligand charge on protein structure. Langmuir 2005, 21, 12080–12084. [Google Scholar] [CrossRef] [PubMed]
- Edwards, R.A.; Whittaker, M.M.; Whittaker, J.W.; Baker, E.N.; Jameson, G.B. Removing a Hydrogen Bond in the Dimer Interface of Escherichia coli Manganese Superoxide Dismutase Alters Structure and Reactivity. Biochemistry 2001, 40, 4622–4632. [Google Scholar] [CrossRef] [PubMed]









| NP:MUA | 1:0 | 1:100 | 1:250 | 1:500 | 1:750 | 1:1000 | rVuFeSOD (Dimer Size) | |
|---|---|---|---|---|---|---|---|---|
| Protein layer thickness * | Physical adsorption | 6.1 | 5.4 | 4.7 | 4.3 | 3.5 | 3.1 | 7.8 |
| Covalent | - | 5.6 | 6.0 | 4.8 | 4.7 | 4.9 | ||
| ∆ λLSPR (nm) | Physical adsorption | 7.0 | 6.8 | 6.5 | 5.6 | 5.0 | 4.3 | - |
| Covalent | - | 7.2 | 7.1 | 7.2 | 7.0 | 6.0 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tellechea, E.; Asensio, A.C.; Ciaurriz, P.; Buezo, J.; López-Gómez, P.; Urra, M.; Moran, J.F. A Study of the Interface of Gold Nanoparticles Conjugated to Cowpea Fe-Superoxide Dismutase. Antioxidants 2022, 11, 2082. https://doi.org/10.3390/antiox11112082
Tellechea E, Asensio AC, Ciaurriz P, Buezo J, López-Gómez P, Urra M, Moran JF. A Study of the Interface of Gold Nanoparticles Conjugated to Cowpea Fe-Superoxide Dismutase. Antioxidants. 2022; 11(11):2082. https://doi.org/10.3390/antiox11112082
Chicago/Turabian StyleTellechea, Edurne, Aaron C. Asensio, Paula Ciaurriz, Javier Buezo, Pedro López-Gómez, Marina Urra, and Jose F. Moran. 2022. "A Study of the Interface of Gold Nanoparticles Conjugated to Cowpea Fe-Superoxide Dismutase" Antioxidants 11, no. 11: 2082. https://doi.org/10.3390/antiox11112082