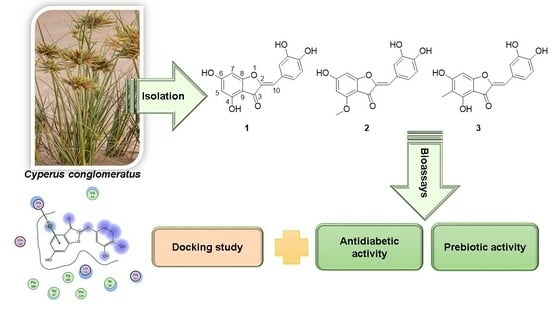
Aurones as Antidiabetic Agents and Their Prebiotic Activities
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Extraction and Isolation
2.3. Assessment of Biological Activities
2.3.1. In Vitro Glycogen Phosphorylase Inhibition Assay
2.3.2. Inhibition of α-Amylase
2.3.3. Inhibition of α-Glucosidase
2.4. Prebiotic Activity
2.4.1. Strains
2.4.2. Prebiotic Activity Score (Pscore)
2.5. Docking Studies
2.5.1. Preparation of the Target
2.5.2. Docking of the Tested Molecules to the Enzyme Binding Site
2.6. Statistical Analysis
3. Results and Discussions
3.1. Identification of Isolated Compounds
3.2. Assessment of Antidiabetic Activity of the Isolated Compounds
3.3. Prebiotic Activity Score of the Isolated Compounds
3.4. Docking Studies
3.4.1. Docking Analysis of the Test Samples and Positive Control (Acarbose) against α-Amylase
3.4.2. Docking Analysis of α-Glucosidase
3.4.3. Docking Analysis of Glycogen Phosphorylase
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cragg, G.M.; Newman, D.J. Natural products: A continuing source of novel drug leads. Biochim. Biophys. Acta (BBA) Gen. Subj. 2013, 1830, 3670–3695. [Google Scholar] [CrossRef] [PubMed]
- Nakib, R.; Ghorab, A.; Ouelhadj, A.; Rodríguez-Flores, S.; Escuredo, O.; Bensouici, C.; Seijo-Coello, C. Chemometric evaluation of antioxidant activity and α-amylase inhibition of selected monofloral honeys from Algeria. J. Apic. Res. 2021, 1–11. [Google Scholar] [CrossRef]
- de Souza, P.M.; de Sales, P.M.; Simeoni, L.A.; Silva, E.C.; Silveira, D.; de Oliveira Magalhães, P. Inhibitory activity of α-amylase and α-glucosidase by plant extracts from the Brazilian cerrado. Planta Med. 2012, 78, 393–399. [Google Scholar] [CrossRef] [PubMed]
- Herowati, R.; Widodo, G.P. Molecular Docking studies of chemical constituents of Tinospora cordifolia on glycogen phosphorylase. Procedia Chem. 2014, 13, 63–68. [Google Scholar] [CrossRef]
- Ajayi, O.S.; Balogun, O.S.; Olawuni, I.J.; October, N.; Adigun, R.; Akinlade, I.G. Alpha Amylase Inhibition and Antioxidant Activities of Bicyclic Diterpenoid Lactones from Andrographis paniculata. Trop. J. Nat. Prod. Res. (TJNPR) 2021, 5, 1110–1117. [Google Scholar] [CrossRef]
- Arraki, K.; Totoson, P.; Decendit, A.; Zedet, A.; Maroilley, J.; Badoc, A.; Demougeot, C.; Girard, C. Mammalian Arginase Inhibitory Activity of Methanolic Extracts and Isolated Compounds from Cyperus Species. Molecules 2021, 26, 1694. [Google Scholar] [CrossRef] [PubMed]
- Jyoti, P.; Hemali, P.; Nilam, R.; Sumitra, C. Cyperus conglomeratus (Cyperaceae) a halophyte from Gujarat: Physicochemical, Phytochemical and Pharmacognostic studies. J. Phytopharmacol. 2018, 7, 334–340. [Google Scholar] [CrossRef]
- Al-Hazmi, G.H.; Awaad, A.S.; Alothman, M.R.; Alqasoumi, S.I. Anticandidal activity of the extract and compounds isolated from Cyperus conglomeratus Rottb. Saudi Pharm. J. 2018, 26, 891–895. [Google Scholar] [CrossRef]
- Hisham, A.; Rameshkumar, K.B.; Sherwani, N.; Al-Saidi, S.; Al-Kindy, S. The composition and antimicrobial activities of Cyperus conglomeratus, Desmos chinensis var. lawii and Cyathocalyx zeylanicus essential oils. Nat. Prod. Commun. 2012, 7, 663–666. [Google Scholar] [CrossRef]
- Al-Harbi, K.B.; El-Ashmawy, I.M.; Al-Wabel, N.A. The antidiarrheal activity of the methanol extract of some plants native to Al-Qassim Region, Saudi Arabia. J. Food Agric. Environ. 2016, 14, 239. [Google Scholar]
- Al-Harbi, K.B.; El-Tigani-Asil, E.; Ahmed, A.F.; El-Ashmawy, I.M.; Al-Wabel, N.A. Wound healing potential of methanolic extracts of some plants native to Al-Qassim Region, Saudi Arabia. J. Food Agric. Environ. 2016, 14, 238–242. [Google Scholar]
- Feizbakhsh, A.; Naeemy, A. Chemical composition of the essential oil of Cyperus conglomeratus Rottb. from Iran. J. Chem. 2011, 8, 241325. [Google Scholar]
- Abdel-Mogib, M.; Basaif, S.; Ezmirly, S. Two novel flavans from Cyperus conglomeratus. Pharmazie 2000, 55, 693–695. [Google Scholar] [PubMed]
- Abdel-Razik, A.F.; Nassar, M.I.; El-Khrisy, E.-D.A.; Dawidar, A.-A.M.; Mabry, T.J. New prenylflavans from Cyperus conglomeratus. Fitoterapia 2005, 76, 762–764. [Google Scholar] [CrossRef] [PubMed]
- Zaki, A.A.; Ross, S.A.; El-Amier, Y.A.; Khan, I.A. New flavans and stilbenes from Cyperus conglomeratus. Phytochem. Lett. 2018, 26, 159–163. [Google Scholar] [CrossRef]
- Haudecoeur, R.; Boumendjel, A. Recent advances in the medicinal chemistry of aurones. Curr. Med. Chem. 2012, 19, 2861–2875. [Google Scholar] [CrossRef]
- Lee, E.H.; Song, D.-G.; Lee, J.Y.; Pan, C.-H.; Um, B.H.; Jung, S.H. Inhibitory effect of the compounds isolated from Rhus verniciflua on aldose reductase and advanced glycation endproducts. Biol. Pharm. Bull. 2008, 31, 1626–1630. [Google Scholar] [CrossRef]
- Dawood, D.H.; Darwish, M.S.; El-Awady, A.A.; Mohamed, A.H.; Zaki, A.A.; Taher, M.A. Chemical characterization of Cassia fistula polysaccharide (CFP) and its potential application as a prebiotic in synbiotic preparation. RSC Adv. 2021, 11, 13329–13340. [Google Scholar] [CrossRef]
- Megur, A.; Daliri, E.B.-M.; Baltriukienė, D.; Burokas, A. Prebiotics as a tool for the prevention and treatment of obesity and diabetes: Classification and ability to modulate the gut microbiota. Int. J. Mol. Sci. 2022, 23, 6097. [Google Scholar] [CrossRef]
- Kobyliak, N.; Virchenko, O.; Falalyeyeva, T. Pathophysiological role of host microbiota in the development of obesity. Nutr. J. 2015, 15, 43. [Google Scholar] [CrossRef]
- Jandhyala, S.M.; Talukdar, R.; Subramanyam, C.; Vuyyuru, H.; Sasikala, M.; Reddy, D.N. Role of the normal gut microbiota. World J. Gastroenterol. WJG 2015, 21, 8787. [Google Scholar] [CrossRef]
- Combettes, M.M. GLP-1 and type 2 diabetes: Physiology and new clinical advances. Curr. Opin. Pharmacol. 2006, 6, 598–605. [Google Scholar] [CrossRef] [PubMed]
- Li, H.-Y.; Zhou, D.-D.; Gan, R.-Y.; Huang, S.-Y.; Zhao, C.-N.; Shang, A.; Xu, X.-Y.; Li, H.-B. Effects and mechanisms of probiotics, prebiotics, synbiotics, and postbiotics on metabolic diseases targeting gut microbiota: A narrative review. Nutrients 2021, 13, 3211. [Google Scholar] [CrossRef]
- Elbermawi, A.; Darwish, M.S.; El-Awady, A.A.; Zaki, A.A.; Qiu, L.; Samra, R.M. Isolation and biological activities of compounds from Rumex vesicarius L. and their use as a component of a synbiotic preparation. Food Chem. X 2022, 14, 100306. [Google Scholar] [CrossRef]
- Elbermawi, A.; Darwish, M.S.; Zaki, A.A.; Abou-Zeid, N.A.; Taher, M.A.; Khojah, E.; Bokhari, S.A.; Soliman, A.F. In Vitro Antidiabetic, Antioxidant, and Prebiotic Activities of the Chemical Compounds Isolated from Guizotia abyssinica. Antioxidants 2022, 11, 2482. [Google Scholar] [CrossRef] [PubMed]
- Abdallah, H.M.; Kashegari, A.T.; Shalabi, A.A.; Darwish, K.M.; El-Halawany, A.M.; Algandaby, M.M.; Ibrahim, S.R.; Mohamed, G.A.; Abdel-Naim, A.B.; Koshak, A.E. Phenolics from Chrozophora oblongifolia Aerial Parts as Inhibitors of α-Glucosidases and Advanced Glycation End Products: In-Vitro Assessment, Molecular Docking and Dynamics Studies. Biology 2022, 11, 762. [Google Scholar] [CrossRef] [PubMed]
- Brayer, G.D.; Luo, Y.; Withers, S.G. The structure of human pancreatic α-amylase at 1.8 Å resolution and comparisons with related enzymes. Protein Sci. 1995, 4, 1730–1742. [Google Scholar] [CrossRef] [PubMed]
- Shirai, T.; Hung, V.S.; Morinaka, K.; Kobayashi, T.; Ito, S. Crystal structure of GH13 α-glucosidase GSJ from one of the deepest sea bacteria. Proteins Struct. Funct. Bioinform. 2008, 73, 126–133. [Google Scholar] [CrossRef]
- Oikonomakos, N.G.; Chrysina, E.D.; Kosmopoulou, M.N.; Leonidas, D.D. Crystal structure of rabbit muscle glycogen phosphorylase a in complex with a potential hypoglycaemic drug at 2.0 Å resolution. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2003, 1647, 325–332. [Google Scholar] [CrossRef]
- Samra, R.M.; Soliman, A.F.; Zaki, A.A.; Ashour, A.; Al-Karmalawy, A.A.; Hassan, M.A.; Zaghloul, A.M. Bioassay-guided isolation of a new cytotoxic ceramide from Cyperus rotundus L. S. Afr. J. Bot. 2021, 139, 210–216. [Google Scholar] [CrossRef]
- Shrestha, S.; Lee, D.-Y.; Park, J.-H.; Cho, J.-G.; Lee, D.-S.; Li, B.; Kim, Y.-C.; Jeon, Y.-J.; Yeon, S.-W.; Baek, N.-I. Flavonoids from the fruits of Nepalese sumac (Rhus parviflora) attenuate glutamate-induced neurotoxicity in HT22 cells. Food Sci. Biotechnol. 2013, 22, 895–902. [Google Scholar] [CrossRef]
- Sayed, H.M.; Mohamed, M.; Farag, S.; Mohamed, G.; Ebel, R.; Omobuwajo, O.; Proksch, P. Phenolics of Cyperus alopecuroides rottb. Inflorescences and their biological activities. Bull. Pharm. Sci. Assiut 2006, 29, 9–32. [Google Scholar] [CrossRef]
- Seabra, R.M.; Silva, A.M.; Andrade, P.B.; Moreira, M.M. Methylaurones from Cyperus capitatus. Phytochemistry 1998, 48, 1429–1432. [Google Scholar] [CrossRef]
- Roshanzamir, K.; Kashani-Amin, E.; Ebrahim-Habibi, A.; Navidpour, L. Aurones as new porcine pancreatic α-Amylase inhibitors. Lett. Drug Des. Discov. 2019, 16, 333–340. [Google Scholar] [CrossRef]
- Sun, H.; Song, X.; Tao, Y.; Li, M.; Yang, K.; Zheng, H.; Jin, Z.; Dodd, R.H.; Pan, G.; Lu, K. Synthesis & α-glucosidase inhibitory & glucose consumption-promoting activities of flavonoid–coumarin hybrids. Future Med. Chem. 2018, 10, 1055–1066. [Google Scholar]




| % Inhibition | |||||
|---|---|---|---|---|---|
| α-Glucosidase | α-Amylase | GP | |||
| Acarbose | 97.7 ± 0.54 | Acarbose | 97.7 ± 0.88 | CP-91149 | 84.9 ± 0.81 |
| 1 | 60.5 ± 1.54 | 1 | 58.4 ± 1.61 | 1 | 27.5 ± 0.64 |
| 2 | 20.91 ± 1.467 | 2 | 39.57 ± 0.592 | 2 | 17.3 ± 1.79 |
| 3 | 79.6 ± 0.97 | 3 | 53.7 ± 1.03 | 3 | 80.4 ± 1.34 |
| Lb. rhamnosus | Lb. paracasei | |
|---|---|---|
| 1 | 3.84 ± 0.67 | 1.82 ± 0.14 |
| 2 | 3.82 ± 0.73 | 3.81 ± 0.56 |
| 3 | 2.36 ± 0.76 | 4.55 ± 0.78 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Samra, R.M.; Darwish, M.S.; Abou-Zeid, N.A.; Khojah, E.; Imieje, V.O.; Zaki, A.A. Aurones as Antidiabetic Agents and Their Prebiotic Activities. Future Pharmacol. 2023, 3, 625-636. https://doi.org/10.3390/futurepharmacol3030040
Samra RM, Darwish MS, Abou-Zeid NA, Khojah E, Imieje VO, Zaki AA. Aurones as Antidiabetic Agents and Their Prebiotic Activities. Future Pharmacology. 2023; 3(3):625-636. https://doi.org/10.3390/futurepharmacol3030040
Chicago/Turabian StyleSamra, Reham M., Mohamed S. Darwish, Noha A. Abou-Zeid, Ebtihal Khojah, Vincent O. Imieje, and Ahmed A. Zaki. 2023. "Aurones as Antidiabetic Agents and Their Prebiotic Activities" Future Pharmacology 3, no. 3: 625-636. https://doi.org/10.3390/futurepharmacol3030040