1. Introduction
The tomato virome consists of at least 136 characterized virus species threatening tomato production worldwide plus a number of new viruses that are being progressively identified by next-generation sequencing [
1]. The role of these not yet characterized viruses in pathogenesis, disease development, and crop losses is unknown. Characterized and economically relevant viruses infecting tomato include the strains of tomato spotted wilt virus (TSWV) breaking the resistance conferred by the SW5 gene (Sw5-RB) (TSWV), strains of cucumber mosaic virus (CMV) harboring harmful variants of their satellite RNAs (satRNA), and isolates of potato virus Y (PVY) necrogenic to tomato, which appeared since the 1980s, and quickly became one of the most important tomato diseases in the Mediterranean countries. TSWV, CMV, and PVY are transmitted by arthropods and are included among the top ten economically important plant viruses [
2] for which efficient and environmentally friendly methods of control are not yet available. Means of natural transmission, extended host range, and appearance of new or recombinant strains with increased virulence may account for control failures. CMV infects approximately 1300 monocots and dicots within 500 genera of more than 100 botanical families [
3] including crop and wild species, which are important for virus persistence in open field [
2,
4]. TSWV host range is also large, consisting of at least 1090 species among crop and wild plants in 84 botanical families [
2,
5,
6]. Finally, PVY has a relatively narrower host range compared to TSWV and CMV but, similar to them, new and emerging strains may cause severe epidemics in pepper, potato, tobacco, and tomato [
2,
7,
8]. In spite of several attempts to obtain suitable levels of resistance by classical breeding or transgenesis [
4,
8,
9,
10,
11], field control of these viruses is still based on routine pesticide sprays against vectors, which proved scantily efficient because of the stylet-borne nature of the non-persistent transmission of CMV and PVY mediated by aphids and the complex persistent propagative relationship between thrips and TSWV, which replicates in both plants and thrips, thus leading to the continuous emergence of resistance-breaking strains and new species [
12]. On the other hand, introgression of potential resistance genes into commercial crops proved ineffective against virus strains with high genome plasticity and superior virulence. Several recessive (sw2, sw3, and sw4) and dominant (Sw1a and Sw1b) resistance genes against TSWV have been reported and incorporated into commercial tomato cultivars (see for review [
9] and references quoted therein) but their resistance was isolate-specific and, therefore, it was quickly overcome by TSWV infection. Only the Sw-5 gene cluster originated from
Solanum peruvianum provided durable and stable resistance against TSWV isolates from different geographic areas. However, large deployment of such Sw5-TSWV-resistant tomato varieties has resulted in the onset of RB isolates overcoming the resistance by single-amino acid substitutions in the TSWV NSM protein [
13,
14]. Resistance sources against CMV have been identified within the genus
Solanum, but all were strain specific and/or quantitative [
15]. Thus, CMV-resistant tomato varieties are not yet commercially available. Transgenic resistance of tomato varieties expressing the CMV coat protein or beneficial satRNAs has been shown to be partially [
16,
17] or highly efficient [
18,
19], respectively, in field trials. However, implementation of satRNA-mediated protection in agriculture is still a matter of debate for potential risks [
20]. Necrogenic strains of PVY induce necrotic lesions on tomato leaves followed by a streak on stems with all the commercial hybrids and varieties commonly grown in greenhouses and open field. Therefore, unlike what is described in potato, pepper, and tobacco, PVY did not reveal any strain specialization to infect tomato [
21]. This, in theory, should have facilitated the search for PVY resistance genes to be introgressed in tomato. According to Parrella et al. [
22], resistance against PVY and another potyvirus tobacco etch virus has been identified in the wild tomato accession “PI 247087.” The resistance gene corresponds to a eukaryotic translation initiation factor that interacts with viral VPg and prevents virus accumulation in inoculated tissues [
23]. In spite of these results, PVY-resistant tomato varieties are not yet available on the market. Thus, introgression of potential resistance genes into commercial crops proved ineffective against virus strains with high genome plasticity and superior virulence.
Therefore, alternative integrated management strategies were exploited.
Suitable levels of resistance to abiotic and biotic stresses have been successfully obtained in vegetable annual crops by grafting two genotypes selected as scion and rootstock. Actually, grafting offers a rapid surgical alternative to the time-consuming classical breeding or transgenesis to combine desired characteristics of rootstock and scion in vegetable crops [
24,
25].
Ideally, scion must cope with crop performance and yield required by farmers and fruit quality and nutritional value required by consumers [
26]. The rootstock should guarantee cultivation under adverse environments such as salinity, nutrient deficiency, drought, pollutants, and soil-borne pathogens such as fungi, oomycetes, bacteria, and viruses [
27,
28,
29,
30,
31,
32,
33]. Among vegetable crops, tomato, eggplant, sweet pepper, watermelon, melon, and cucumber are commonly and economically grafted in Asia, Europe, and North America. To give some examples (reviwed by [
34]), watermelon, melon, and cucumber are prevalently grafted onto
Cucurbita maxima ×
C. moschata rootstocks such as “Shintoza,” “Ps1313,” “TZ 148” or onto
C. moschata “Jinxinzhen,”
C. pepo “Brava,” and
C. ficifolia in the case of cucumber. Tomato has been grafted mainly onto tomato genotypes and interspecific hybrids such as the
Solanum lycopersicum ×
S. habrochaites “Maxifort” and “Beaufort” rootstocks, which improve crop yield, tolerance to soilborne pathogens and fruit quality, favoring higher accumulation of phenolic compounds, vitamin C, lycopene, and flavonoids in fruits of grafted plants [
35]. Eggplant has been grafted traditionally on
Solanum spp. wild species such as
S. integrifolium,
S. torvum, and
S. sisymbriifolium, which provide high level of resistance to bacterial wilt, Fusarium wilt, Verticillium wilt, and root-knot nematodes. Compared to tomato and eggplant, grafted sweet peppers are not typically commercialized. Fruit shape is important in pepper crops but certain rootstocks can modulate gene expression and fruit development of the scion. Tsaballa et al. [
36] demonstrated that phenotypic change in sweet pepper can be inherited for up to two generations of seed produced by these progenies. Nonetheless, suitable levels of resistance against root-knot nematodes and
Phytophthora capsici were attained in sweet pepper grafted on
Capsicum annuum accessions “AR96023” and “AF2638,” respectively. Finally, this list could not be exhaustive without including the control of Verticillium wilts obtained by grafting seed-propagated globe artichoke hybrids onto cardoon [
37] as well as the list of potential rootstocks with special characteristics to manage biotic and abiotic stresses in tomato, eggplant, chili, potato, cucumber, muskmelon, pumpkin, and wax gourd reported by Kumar et al. [
38].
Grafted tomato plants were implemented in the early 1960s but their use aroused little interest because farmers were particularly discouraged by the higher price of grafted plants compared to non-grafted seedlings. Recent surveys report that the price of grafted tomato seedlings for fresh market tomato production is between 0.4 to 1.2 USD per grafted seedling in the USA and some Asian countries, including Japan and Korea, and between 0.6 to 1.2 EUR per grafted seedling in Spain and some European countries [
39]. Nonetheless, actual estimates suggest between 20% and 40% of tomatoes are grafted [
39] and their implementation is rapidly increasing all over the world in organic and environmentally friendly crops for the control of soil-borne pathogens as an alternative to the banned methyl bromide and to mitigate effects of abiotic stresses.
The higher cost of the grafted plantlets is now compensated by a number of advantages, which include production increase and earliness of the harvest with a reduced number of plants, rational use of irrigation water, use of fewer fertilizers, and elongation of the crop cycle. A good rootstock/scion combination usually guarantees a robust root system and the maintenance of good vegetative vigor and resistance to deal with abiotic and biotic stresses until the end of the farming cycle [
27]. All these aspects guarantee net returns large enough to make grafted tomatoes significantly more profitable than non-grafted seedlings [
40]. Grafting is a viable approach also to limit damage by viruses transmitted through soil such as melon necrotic spot virus (MNSV) vectored by
Olpidium sp. or the contact-transmissible tobacco mosaic virus (TMV), which reaches the soil in infected plant debris or as seed-coat contaminant. Rootstocks with specific characteristics have been developed and deployed for the control of MNSV in muskmelon and watermelon [
41,
42,
43] and of TMV in tomato [
38]. However, the majority of plant viruses is airborne by arthropods and pollen or transmitted by contact or aerosol [
44,
45] (
Table 1).
Some promising attempts were recently made to reduce incidence of tomato yellow leaf curl virus, TSWV, and pepino mosaic virus in different scion-rootstocks combinations [
46,
47,
48,
49].
2. Why Grafting?
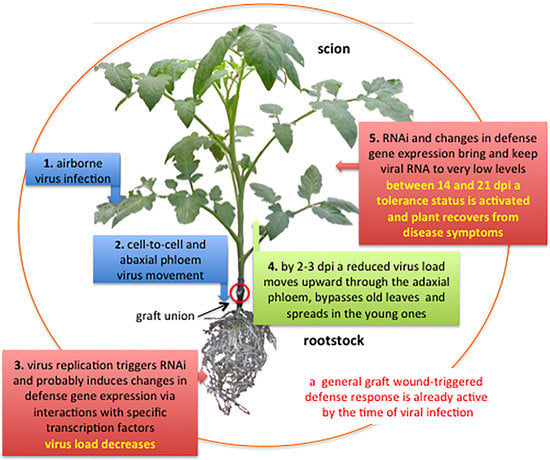
Samuel [
50] described a general pattern of translocation of tobacco mosaic virus (TMV) into plant tissues suggesting that after entering tobacco leaf tissue, TMV particles primarily reach roots through phloem sieve tubes and between 2 to 3 days post-inoculation (dpi) they follow phloem sap in the reverse way up, bypass the old leaves, and invade systemically the young plant tissues [
51]. Andrianifahanana et al. [
52] reported a similar pattern of virus translocation in pepper of the potyvirus pepper mottle virus. The authors documented virus movement from the inoculated leaf down the stem toward the roots via the external phloem. Then the virus entered internal phloem at the cotyledon node and rapidly spread upward to the young tissues. A phloem-translocation pattern has been described also for cauliflower mosaic virus [
53], cherry leafroll virus [
54], and TMV [
55] (reviewed in [
56,
57,
58]). Internal and external phloem is present in the stem of
Solanaceae and of some other plant families such as
Cucurbitaceae and corresponds to the adaxial and abaxial phloem strands of leaf veins and petioles. In tomato leaflets the xylem is in the center of the vein with the phloem distributed on both the adaxial and abaxial sides of the bundle. The abaxial phloem exports sugars from the leaf in a descending direction to roots. Thus, a virus entering abaxial leaf phloem is expected to follow transport of photoassimilates down to roots, according to the source-sink relationship [
59]. For potyviruses, virus loading in the external phloem depends on the virus-host combination [
56] and for potato virus A it is controlled by the virus-coded Vpg [
60]. On the contrary, a complex structure of CMV containing viral RNA, coat protein, and the 3a movement protein loads into sieve elements of minor veins of
Nicotiana clevelandii. The complex successively assembles into complete virus particles within minor vein sieve elements [
58,
61]. Unfortunately no such data are available for TSWV.
Results from these studies were seminal to our idea to exploit grafting as a strategy to control airborne viral infections in tomato.
This review gives a brief account of the results and proposes mechanisms probably involved in the resistance/tolerance observed in grafted plants of the three cases-study carried out in our laboratory to manage one Sw5-RB strain of TSWV [
62], two CMV strains supporting stunting and necrogenic satRNAs [
63], and one recombinant PVY strain necrogenic to tomato [
64].
3. Screening of Solanum spp. Germplasm
Local tomato ecotypes and commercial hybrids and Solanum spp. genotypes were screened for resistance/tolerance to challenge inoculation of specific strains of TSWV, CMV, and PVY characterized by superior virulence.
Screening for potential rootstocks privileged local tomato ecotypes found and characterized in the framework of regional actions aimed at preserving biodiversity of vegetable crops in Apulia (southern Italy). Selected ecotypes were Fiaschetto (Fi), Giallo invernale (Gi), Manduria (Ma), Morciano (Mor), Racalino (R), and Regina (Re). Screenings included also
Solanum integrifolium,
S. nigrum,
S. torvum, eggplant cv. Molfettese (Mo), and commercial tomato hybrids, Faino (Fa), Messapico (Me), Taylor (Ta) or not Pullrex (Pu), carrying the Sw5-resistance gene to TSWV and the tomato variety UC82 (UC) as highly susceptible control. Tests were carried out with cleft-grafted and non-grafted plants rub-inoculated on the first leaf above the graft junction with sap extracted from infected plants and grown under glasshouse [
62,
63,
64] or exposed to field inoculum (this review). According to Kumar et al. [
38], complete repair of graft wound would be achieved between 7 and 10 days after grafting, thus we were confident that by the time of inoculation, continuous cambial connections between scion and rootstock had been restored. In glasshouse tests, plants were grown and maintained at 24 ± 2 °C with 16 h photoperiod and monitored daily for disease symptoms. Accumulation of viral RNA was estimated by dot blot or tissue print hybridization in samples collected from inoculated and mock-inoculated plants at 14, 21, and 28 days post-inoculation (dpi). Plant tissues were ground in the presence of an alkaline solution and spotted onto positively charged nylon membranes that were hybridized overnight with virus-specific DIG-labeled RNA or DNA probes. Chemiluminescent hybridization signals were used to detect and quantify the accumulation of viral RNA by using
Glyceraldehyde 3-phosphate dehydrogenase (
GAPDH) as housekeeping gene for normalization [
65]. Viral genomic RNAs and virus-specific small interfering RNAs were separated on agarose or polyacrylamide gels, respectively, and detected by northern blot hybridization with specific radiolabeled or DIG-labeled probes [
62,
63] or by NGS sequencing [
64].
For field tests, three biological replicates of each plant at first branching stage were transplanted according to a fully randomized block scheme. In 2016 and 2017, two experimental fields were set up between March and April using 270 and 180 plants per field, respectively. Data were collected before the middle of August, at the end of vegetative season.
3.1. Results with TSWV
S. nigrum and UC showed systemic symptoms ranging from severe mosaic, to leaf and stem necrosis and plant death in response to infection of the Sw5-RB strain TSWV-CiPz, whereas Ma, Mo, and
S. integrifolium showed mild mosaic and, more interestingly, recovered from viral disease symptoms between 21 and 28 dpi (
Table 2). The lowest levels of viral RNA were detected in Ma, Mo, and
S. integrifolium, therefore these genotypes were selected as rootstocks to prepare graft combinations. Self-grafted UC/UC, Ma/Ma, Fa/Fa, Me/Me, and the Me and Fa scions grafted onto Ma showed mild mosaic. All these plants recovered from disease symptoms and displayed increased growth, abundant leaf canopy, and root development, compared to non-grafted infected plants. Oneweek later, also UC/Mo, and Pu/Pu recovered from disease symptoms. Recovery was not observed with non-grafted UC and Pu. Viral RNA load was markedly reduced in the Me/Ma graft combination as well as in all scions of the self-grafted combinations (
Table 2).
In a field test set up in 2016, the only virus detected was a Sw5-RB TSWV strain identified by targeting NS
m sequences flanking the
MaeI restriction site with RT-PCR RFLP [
66] and rub-inoculation onto the commercial tomato hybrids Diaz and York carrying the Sw5 resistance gene [
67]. Field infection damaged 7–8% of the plants with necrotic symptoms on leaves and fruits. Among the tomato ecotypes evaluated, Regina (Re) proved the most susceptible (
Figure 1) but despite the overall low incidence of viral infection in the field plants self-grafted or grafted onto Ma showed a productive advantage compared to the non-grafted counterparts. In a second field test set up in 2017, we exposed to field inoculum the Sw5 commercial tomato hybrid Taylor self-grafted or grafted onto Ma. Non-grafted Taylor plants served as control. Again, the only virus detected was an Sw5-RB TSWV strain. Graft combinations Taylor/Taylor and Taylor/Ma produced between 6 and 9 kg/plant, whereas the non-grafted Taylor counterparts produced between 4 and 5 kg/plant, which is thought to be the standard production of this hybrid. In addition, the non-grafted Taylor plants, showed sporadic symptoms of Sw5-RB TSWV infection and about ten days delay in fruit ripening, compared to the grafted plants.
3.2. Results with CMV
CMV infections in tomato crops became suddenly relevant around the 1980s after the introduction of the so-called “Asian” strains characterized by superior aggressiveness compared to endemic CMV strains [
68]. After initial severe outbreaks between 1988 and 1992, CMV has been detected sporadically in tomato fields although outbreaks may still occur after very mild winters, which may lead to huge increases in overwintering aphid populations that spread the virus from infected foci in wild plants communities into the crops [
68,
69]. Therefore we limited our study to glasshouse tests. Infection of CMV-TTS carrying a tomato top stunting satRNA induced leaf distortion and apical stunting in UC and Ma (
Table 3), which persisted in UC until 30 dpi but not in Ma plants that recovered from disease symptoms. Symptoms elicited in non-grafted plants were very similar to those of grafted plants but, by 21 dpi, the latter fully recovered from disease symptoms (
Table 3). Viral RNA loads in Ma were about 1.6-fold lower than in UC whereas no significative differences were estimated among grafted and self-grafted plants. Systemic infection of CMV-77 carrying a necrogenic satRNA induced stunting and severe stem and leaf necrosis in both Ma and UC, between 15 and 18 dpi (
Table 3). Ma plants transiently recovered from necrosis between 18 and 21 dpi but by 30 dpi, these plants also died (
Table 3). Non-grafted genotypes showed a more severe and rapid disease progression than grafted plants. Necrosis developed in all UC self-grafted plants and in one out of the four UC/Ma plants but none of the plants died. Interestingly, self-grafted Ma plants recovered from necrosis by 21 dpi (
Table 3). Because of the occurrence of a necrotic phenotype, we monitored the accumulation of viral RNA at 15 dpi. At this time-point, viral RNA load in Ma was 8-fold lower than in UC and this ratio was substantially similar among the three graft combinations tested (
Table 3).
3.3. Results with PVY
PVY isolates infecting tomato show high genetic diversity, including genetic recombination. One of such recombinants, denoted PVY
C-to, was characterized as an interlineage recombinant isolate of the PVY
C group and associated with necrotic spots and vein necrosis on the leaflets and pale yellow spots scattered on fruit skin of a table tomato variety [
70].
In two independent glasshouse experiments, PVY
C-to infected systemically UC and Ma. UC infection was characterized by mosaic, leaf blade reduction, and twisting with some necrotic spots scattered on the leaf surface. Conversely, infection in Ma was substantially asymptomatic but viral RNA loads at 14 dpi did not differ significantly between UC and Ma, despite the differences in symptoms. Infection of PVY
C-to in the three graft combinations induced a mild distortion of leaf margin and a slightly reduced growth. Viral RNA loads at 14 dpi did not differ significantly among the three graft combinations but was approximately 2.5-fold higher than in non-grafted plants. Despite the difference in viral RNA loads, non-grafted Ma and all grafted plants recovered from disease symptoms by 21 dpi with a mean of 3-fold reduction in viral RNA loads compared with the estimates at 14 dpi sampling time. Non-grafted UC plants did not recover from disease symptoms; rather, they suffered increased disease severity and a further 2-fold increase in viral RNA loads by passing from 14 to 21 dpi (
Table 4).