Functional Imaging and Inhibitor Screening of Human Pancreatic Lipase by a Resorufin-Based Fluorescent Probe
Abstract
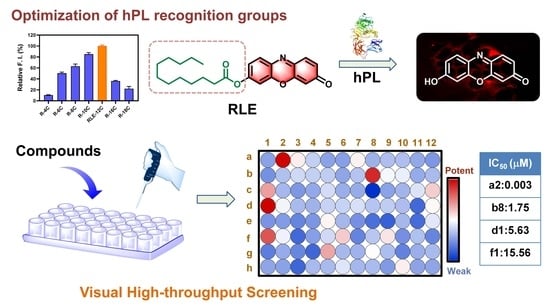
:1. Introduction
2. Materials and Methods
2.1. Materials and Instruments
2.2. Chemistry
2.3. Selectivity and Sensitivity of RLE to hPL
2.4. Enzymatic Kinetics of hPL-Mediated RLE Hydrolysis
2.5. Cell Viability Assay
2.6. Confocal Microscopic Imaging of PL in AR42J Cells
2.7. Fluorescence Imaging of Monkey Pancreas Slices
2.8. Molecular Docking Simulations
2.9. hPL Inhibition Assay
3. Results and Discussion
3.1. Design, Synthesis and Sensing Mechanisms of RLE
3.2. Selectivity and Sensitivity of RLE towards hPL
3.3. Enzymatic Kinetics of RLE Hydrolysis
3.4. Confocal Microscopic Imaging of PL in Living Cells and Tissue Slices
3.5. High-Throughput Screening of hPL Inhibitors Based on RLE
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kumar, A.; Chauhan, S. Pancreatic lipase inhibitors: The road voyaged and successes. Life Sci. 2021, 271, 119115. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, M. Human digestive and metabolic lipases—A brief review. J. Mol. Catal. B-Enzym. 2003, 22, 369–376. [Google Scholar] [CrossRef]
- Whitcomb, D.C.; Lowe, M.E. Human pancreatic digestive enzymes. Dig. Dis. Sci. 2007, 52, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Lowe, M.E. Pancreatic triglyceride lipase and colipase: Insights into dietary fat digestion. Gastroenterology 1994, 107, 1524–1536. [Google Scholar] [CrossRef]
- Dikaiou, P.; Björck, L.; Adiels, M.; Lundberg, C.E.; Mandalenakis, Z.; Manhem, K.; Rosengren, A. Obesity, overweight and risk for cardiovascular disease and mortality in young women. Eur. J. Prev. Cardiol. 2021, 28, 1351–1359. [Google Scholar] [CrossRef]
- Cleven, L.; Krell-Roesch, J.; Nigg, C.R.; Woll, A. The association between physical activity with incident obesity, coronary heart disease, diabetes and hypertension in adults: A systematic review of longitudinal studies published after 2012. BMC Public Health 2020, 20, 726. [Google Scholar] [CrossRef]
- Seravalle, G.; Grassi, G. Obesity and hypertension. Pharmacol. Res. 2017, 122, 1–7. [Google Scholar] [CrossRef]
- Sattar, N.; McMurray JJ, V.; McInnes, I.B.; Aroda, V.R.; Lean, M.E.J. Treating chronic diseases without tackling excess adiposity promotes multimorbidity. Lancet Diabetes Endocrinol. 2023, 11, 58–62. [Google Scholar] [CrossRef]
- Heck, A.M.; Yanovski, J.A.; Calis, K.A. Orlistat, a new lipase inhibitor for the management of obesity. Pharmacotherapy 2000, 20, 270–279. [Google Scholar] [CrossRef]
- Filippatos, T.D.; Derdemezis, C.S.; Gazi, I.F.; Nakou, E.S.; Mikhailidis, D.P.; Elisaf, M.S. Orlistat-associated adverse effects and drug interactions: A critical review. Drug Saf. 2008, 31, 53–65. [Google Scholar] [CrossRef]
- Bersoux, S.; Byun, T.H.; Chaliki, S.S.; Poole, K.G. Pharmacotherapy for obesity: What you need to know. Clevel. Clin. J. Med. 2017, 84, 951–958. [Google Scholar] [CrossRef] [PubMed]
- Sanh, N.; Fadul, H.; Hussein, N.; Lyn-Cook, B.D.; Hammons, G.; Ramos-Cardona, X.E.; Mohamed, K.; Mohammed, S.I. Proteomics Profiling of Pancreatic Cancer and Pancreatitis for Biomarkers Discovery. J. Cell Sci. Ther. 2018, 9, 287. [Google Scholar] [PubMed]
- Lasher, D.; Szabó, A.; Masamune, A.; Chen, J.M.; Xiao, X.; Whitcomb, D.C.; Barmada, M.M.; Ewers, M.; Ruffert, C.; Paliwal, S.; et al. Protease-Sensitive Pancreatic Lipase Variants Are Associated With Early Onset Chronic Pancreatitis. Am. J. Gastroenterol. 2019, 114, 974–983. [Google Scholar] [CrossRef] [PubMed]
- Jian, J.Y.; Yuan, J.M.; Fan, Y.; Wang, J.C.; Zhang, T.T.; Jeroen, K.; Zhengjin, J. High-Resolution Bioassay Profiling with Complemented Sensitivity and Resolution for Pancreatic Lipase Inhibitor Screening. Molecules 2022, 27, 6923. [Google Scholar] [CrossRef] [PubMed]
- Moharana, T.R.; Rao, N.M. Substrate structure and computation guided engineering of a lipase for omega-3 fatty acid selectivity. PLoS ONE 2020, 15, e0231177. [Google Scholar] [CrossRef] [Green Version]
- Jin, Q.; Feng, L.; Wang, D.D.; Dai, Z.R.; Wang, P.; Zou, L.W.; Liu, Z.H.; Wang, J.Y.; Yu, Y.; Ge, G.B.; et al. A Two-Photon Ratiometric Fluorescent Probe for Imaging Carboxylesterase 2 in Living Cells and Tissues. ACS Appl. Mater. Interfaces 2015, 7, 28474–28481. [Google Scholar] [CrossRef]
- Wang, P.; Xia, Y.L.; Zou, L.W.; Qian, X.K.; Dou, T.Y.; Jin, Q.; Li, S.Y.; Yu, Y.; Wang, D.D.; Luo, Q.; et al. An Optimized Two-Photon Fluorescent Probe for Biological Sensing and Imaging of Catechol-O-Methyltransferase. Chemistry 2017, 23, 10800–10807. [Google Scholar] [CrossRef]
- Fan, Y.F.; Wu, Y.; Hou, J.; Wang, P.; Peng, X.J.; Ge, G.B. Coumarin-based near-infrared fluorogenic probes: Recent advances, challenges and future perspectives. Coord. Chem. Rev. 2023, 480, 215020. [Google Scholar] [CrossRef]
- Ge, G.B.; Feng, L.; Jin, Q.; Wang, Y.R.; Liu, Z.M.; Zhu, X.Y.; Wang, P.; Hou, J.; Cui, J.N.; Yang, L. A novel substrate-inspired fluorescent probe to monitor native albumin in human plasma and living cells. Anal. Chim. Acta 2017, 989, 71–79. [Google Scholar] [CrossRef]
- Shi, J.; Deng, Q.C.; Wan, C.Y.; Zheng, M.M.; Huang, F.H.; Tang, B. Fluorometric probing of the lipase level as acute pancreatitis biomarkers based on interfacially controlled aggregation-induced emission (AIE). Chem. Sci. 2017, 8, 6188–6195. [Google Scholar] [CrossRef] [Green Version]
- Jacks, T.J.; Kircher, H.W. Fluorometric assay for the hydrolytic activity of lipase using fatty acyl esters of 4-methylumbelliferone. Anal. Biochem. 1967, 21, 279–285. [Google Scholar] [CrossRef] [PubMed]
- Ivanov Sergey, A.; Nomura, K.; Malfanov Ilya, L.; Sklyar Ilya, V.; Ptitsyn Leonid, R. Isolation of a novel catechin from Bergenia rhizomes that has pronounced lipase-inhibiting and antioxidative properties. Fitoterapia 2011, 82, 212–218. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Zhang, S.; Zhen, M.M.; Deng, Q.C.; Zheng, C.; Jing, L.; Huang, F.H. A novel fluorometric turn-on assay for lipase activity based on an aggregation-induced emission (AIE) luminogen. Sens. Actuator Chem. 2017, 238, 765–771. [Google Scholar] [CrossRef] [Green Version]
- Guan, P.; Liu, Y.; Yang, B.; Wu, Y.; Chai, J.; Wen, G.; Liu, B. Fluorometric probe for the lipase level: Design, mechanism and biological imaging application. Talanta 2021, 225, 121948. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Cao, P.; Fan, Z.; Luo, X.; Yang, G.; Qu, T.; Gao, J. Rapid Screening of Lipase Inhibitors in Scutellariabaicalensis by Using Porcine Pancreatic Lipase Immobilized on Magnetic Core-Shell Metal-Organic Frameworks. Molecules 2022, 27, 3475. [Google Scholar] [CrossRef]
- Al-Maharik, N.; Jaradat, N.; Bassalat, N.; Hawash, M.; Zaid, H. Isolation, Identification and Pharmacological Effects of Mandragora autumnalis Fruit Flavonoids Fraction. Molecules 2022, 27, 1046. [Google Scholar] [CrossRef]
- Ma, L.J.; Hou, X.D.; Qin, X.Y.; He, R.J.; Yu, H.N.; Hu, Q.; Guan, X.Q.; Jia, S.N.; Hou, J.; Lei, T.; et al. Discovery of human pancreatic lipase inhibitors from root of Rhodiola crenulata via integrating bioactivity-guided fractionation, chemical profiling and biochemical assay. J. Pharm. Anal. 2022, 12, 683–691. [Google Scholar] [CrossRef]
- Point, V.; Pavan Kumar, K.V.; Marc, S.; Delorme, V.; Parsiegla, G.; Amara, S.; Carrière, F.; Buono, G.; Fotiadu, F.; Canaan, S.; et al. Analysis of the discriminative inhibition of mammalian digestive lipases by 3-phenyl substituted 1,3,4-oxadiazol-2(3H)-ones. Eur. J. Med. Chem. 2012, 58, 452–463. [Google Scholar] [CrossRef]
- Yadav, A.K.; Reinhardt, C.J.; Arango, A.S.; Huff, H.C.; Dong, L.; Malkowski, M.G.; Das, A.; Tajkhorshid, E.; Chan, J. An Activity-Based Sensing Approach for the Detection of Cyclooxygenase-2 in Live Cells. Angew. Chem. 2020, 59, 3307–3314. [Google Scholar] [CrossRef]
- Tian, L.; Feng, H.; Dai, Z.; Zhang, R. Resorufin-based responsive probes for fluorescence and colorimetric analysis. J. Mater. Chem. 2021, 9, 53–79. [Google Scholar] [CrossRef]
- Hu, Q.; Tian, Z.; Wang, H.; Huang, J.; Wang, F.; Zhao, B.; He, R.; Jin, Q.; Hou, X.; Hou, J.; et al. Rational design and development of a novel and highly specific near-infrared fluorogenic substrate for sensing and imaging of human pancreatic lipase in living systems. Sens. Actuators Chem. 2021, 341, 130033. [Google Scholar] [CrossRef]
- Qiao, Z.; Zhang, H.; Zhang, Y.; Wang, K. Detection of Lipase Activity in Cells by a Fluorescent Probe Based on Formation of Self-Assembled Micelles. iScience 2020, 23, 101294. [Google Scholar] [CrossRef] [PubMed]
- Winkler, F.K.; D’Arcy, A.; Hunziker, W. Structure of human pancreatic lipase. Nature 1990, 343, 771–774. [Google Scholar] [CrossRef] [PubMed]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Wu, X.; Shi, W.; Li, X.; Ma, H. Recognition Moieties of Small Molecular Fluorescent Probes for Bioimaging of Enzymes. Acc. Chem. Res. 2019, 52, 1892–1904. [Google Scholar] [CrossRef]
- Poreba, M.; Solberg, R.; Rut, W.; Lunde, N.N.; Kasperkiewicz, P.; Snipas, S.J.; Mihelic, M.; Turk, D.; Turk, B.; Salvesen, G.S.; et al. Counter Selection Substrate Library Strategy for Developing Specific Protease Substrates and Probes. Cell Chem. Biol. 2016, 23, 1023–1035. [Google Scholar] [CrossRef] [Green Version]
- Nagar, S.; Argikar, U.A.; Tweedie, D.J. Enzyme kinetics in drug metabolism: Fundamentals and applications. Methods Mol. Biol. 2014, 1113, 1–6. [Google Scholar]
- Hou, X.D.; Qin, X.Y.; Hou, J.; Tang, H.; Ge, G.B. The potential of natural sources for pancreatic lipase inhibitors: A solution of the obesity crisis? Expert Opin. Drug Discov. 2022, 17, 1295–1298. [Google Scholar] [CrossRef]







Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hou, F.-B.; Zhang, N.; Zhu, G.-H.; Fan, Y.-F.; Sun, M.-R.; Nie, L.-L.; Ge, G.-B.; Zheng, Y.-J.; Wang, P. Functional Imaging and Inhibitor Screening of Human Pancreatic Lipase by a Resorufin-Based Fluorescent Probe. Biosensors 2023, 13, 283. https://doi.org/10.3390/bios13020283
Hou F-B, Zhang N, Zhu G-H, Fan Y-F, Sun M-R, Nie L-L, Ge G-B, Zheng Y-J, Wang P. Functional Imaging and Inhibitor Screening of Human Pancreatic Lipase by a Resorufin-Based Fluorescent Probe. Biosensors. 2023; 13(2):283. https://doi.org/10.3390/bios13020283
Chicago/Turabian StyleHou, Fan-Bin, Na Zhang, Guang-Hao Zhu, Yu-Fan Fan, Meng-Ru Sun, Liang-Liang Nie, Guang-Bo Ge, Yue-Juan Zheng, and Ping Wang. 2023. "Functional Imaging and Inhibitor Screening of Human Pancreatic Lipase by a Resorufin-Based Fluorescent Probe" Biosensors 13, no. 2: 283. https://doi.org/10.3390/bios13020283