Updated Perspectives on Direct Vascular Cellular Reprogramming and Their Potential Applications in Tissue Engineered Vascular Grafts
Abstract
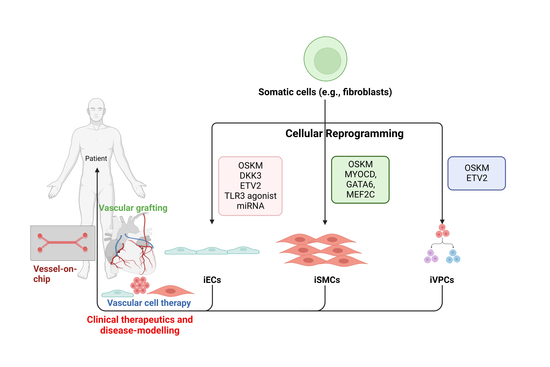
:1. Introduction
2. Endothelial Cell Generation
2.1. Pluripotency Factor-Based Reprogramming
2.2. Lineage-Specific Transcription Factors—The Advent of ETV2
2.3. Innate-Immune Activation—The Big Potential of Small Molecules
2.4. MicroRNA-Based Reprogramming
3. Smooth Muscle Generation
4. Vascular Progenitor Cells
4.1. Pluripotency Factor-Based Reprogramming
4.2. ETV2
5. Tissue Engineered Vascular Grafts
5.1. Decellularised Tissue
5.2. 3D Bioprinting
5.3. Scaffold-Based Grafts
6. The Ethics of Direct Reprogramming
7. Current Challenges & Future Perspectives
7.1. Factor Identification and Reprogramming Efficacies
7.2. Heterogeneity of Derived Cell Populations
7.3. Factor Delivery Systems & Viral Integration
7.4. Tumorigenicity Risk
7.5. Recapitulating Disease
7.6. 2D In Vitro Analysis vs. 3D Microenvironments
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Roth, G.A.; Mensah, G.A.; Johnson, C.O.; Addolorato, G.; Ammirati, E.; Baddour, L.M.; Barengo, N.C.; Beaton, A.Z.; Benjamin, E.J.; Benziger, C.P.; et al. Global Burden of Cardiovascular Diseases and Risk Factors, 1990–2019: Update From the GBD 2019 Study. J. Am. Coll. Cardiol. 2020, 76, 2982–3021. [Google Scholar] [CrossRef] [PubMed]
- McKavanagh, P.; Yanagawa, B.; Zawadowski, G.; Cheema, A. Management and Prevention of Saphenous Vein Graft Failure: A Review. Cardiol. Ther. 2017, 6, 203–223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- SabikIII, J.F. Understanding Saphenous Vein Graft Patency. Circulation 2011, 124, 273–275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pashneh-Tala, S.; MacNeil, S.; Claeyssens, F. The tissue-engineered vascular graft—Past, present, and future. Tissue Eng. Part B Rev. 2016, 22, 68–100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Yang, Y.; Liu, J.; Qian, L. Direct cell reprogramming: Approaches, mechanisms and progress. Nat. Rev. Mol. Cell Biol. 2021, 22, 410–424. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; Kim, J.E.; AL Johnson, B.; Andukuri, A.; Yoon, Y.-S. Direct reprogramming into endothelial cells: A new source for vascular regeneration. Regen. Med. 2017, 12, 317–320. [Google Scholar] [CrossRef] [Green Version]
- Kurian, L.; Sancho-Martinez, I.; Nivet, E.; Aguirre, A.; Moon, K.; Pendaries, C.; Volle-Challier, C.; Bono, F.; Herbert, J.-M.; Pulecio, J.; et al. Conversion of human fibroblasts to angioblast-like progenitor cells. Nat. Methods 2013, 10, 77–83. [Google Scholar] [CrossRef] [Green Version]
- Sayed, N.; Wong, W.T.; Ospino, F.; Meng, S.; Lee, J.; Jha, A.; Dexheimer, P.; Aronow, B.J.; Cooke, J.P. Transdifferentiation of human fibroblasts to endothelial cells role of innate immunity. Circulation 2015, 131, 300–309. [Google Scholar] [CrossRef] [Green Version]
- Margariti, A.; Winkler, B.; Karamariti, E.; Zampetaki, A.; Tsai, T.-N.; Baban, D.; Ragoussis, J.; Huang, Y.; Han, J.-D.J.; Zeng, L.; et al. Direct reprogramming of fibroblasts into endothelial cells capable of angiogenesis and reendothelialization in tissue-engineered vessels. Proc. Natl. Acad. Sci. USA 2012, 109, 13793–13798. [Google Scholar] [CrossRef]
- Li, J.; Huang, N.F.; Zou, J.; Laurent, T.J.; Lee, J.C.; Okogbaa, J.; Cooke, J.P.; Ding, S. Conversion of Human Fibroblasts to Functional Endothelial Cells by Defined Factors. Arter. Thromb. Vasc. Biol. 2013, 33, 1366–1375. [Google Scholar] [CrossRef] [Green Version]
- Hong, X.; Margariti, A.; Le Bras, A.; Jacquet, L.; Kong, W.; Hu, Y.; Xu, Q. Transdifferentiated Human Vascular Smooth Muscle Cells are a New Potential Cell Source for Endothelial Regeneration. Sci. Rep. 2017, 7, 5590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, T.; Karamariti, E.; Hong, X.; Deng, J.; Wu, Y.; Gu, W.; Simpson, R.; Wong, M.M.; Yu, B.; Hu, Y.; et al. DKK3 (Dikkopf-3) Transdifferentiates Fibroblasts Into Functional Endothelial Cells—Brief Report. Arter. Thromb. Vasc. Biol. 2019, 39, 765–773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ginsberg, M.; James, D.; Ding, B.-S.; Nolan, D.; Geng, F.; Butler, J.M.; Schachterle, W.; Pulijaal, V.R.; Mathew, S.; Chasen, S.T.; et al. Efficient Direct Reprogramming of Mature Amniotic Cells into Endothelial Cells by ETS Factors and TGFβ Suppression. Cell 2012, 151, 559–575. [Google Scholar] [CrossRef] [Green Version]
- Wong, W.T.J.; Cooke, J.P. Therapeutic transdifferentiation of human fibroblasts into endothelial cells using forced expression of lineage-specific transcription factors. J. Tissue Eng. 2016, 7, 2041731416628329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.-J.; Kim, D.-H.; Lee, J.Y.; Lee, B.-C.; Kang, I.; Kook, M.G.; Kong, D.; Choi, S.W.; Woo, H.-M.; Kim, D.-I.; et al. cAMP/EPAC Signaling Enables ETV2 to Induce Endothelial Cells with High Angiogenesis Potential. Mol. Ther. 2020, 28, 466–478. [Google Scholar] [CrossRef] [PubMed]
- Morita, R.; Suzuki, M.; Kasahara, H.; Shimizu, N.; Shichita, T.; Sekiya, T.; Kimura, A.; Sasaki, K.-I.; Yasukawa, H.; Yoshimura, A. ETS transcription factor ETV2 directly converts human fibroblasts into functional endothelial cells. Proc. Natl. Acad. Sci. USA 2015, 112, 160–165. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; Park, C.; Han, J.W.; Kim, J.Y.; Cho, K.; Kim, E.J.; Kim, S.; Lee, S.-J.; Oh, S.Y.; Tanaka, Y.; et al. Direct Reprogramming of Human Dermal Fibroblasts Into Endothelial Cells Using ER71/ETV2. Circ. Res. 2017, 120, 848–861. [Google Scholar] [CrossRef] [Green Version]
- McCoy, M.G.; Pérez-Cremades, D.; Belkin, N.; Peng, W.; Zhang, B.; Chen, J.; Sachan, M.; Wara, A.K.M.K.; Zhuang, R.; Cheng, H.S.; et al. A miRNA cassette reprograms smooth muscle cells into endothelial cells. FASEB J. 2022, 36, e22239. [Google Scholar] [CrossRef]
- Yin, L.; Ohanyan, V.; Pung, Y.F.; DeLucia, A.; Bailey, E.; Enrick, M.; Stevanov, K.; Kolz, C.L.; Guarini, G.; Chilian, W.M. Induction of Vascular Progenitor Cells From Endothelial Cells Stimulates Coronary Collateral Growth. Circ. Res. 2012, 110, 241–252. [Google Scholar] [CrossRef]
- Wasteson, P.; Johansson, B.R.; Jukkola, T.; Breuer, S.; Akyürek, L.M.; Partanen, J.; Lindahl, P. Developmental origin of smooth muscle cells in the descending aorta in mice. Development 2008, 135, 1823–1832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bray, S.J. Notch signalling: A simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 2006, 7, 678–689. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Gu, W.; Zhang, G.; Guo, X. Notch1 activation of Jagged1 contributes to differentiation of mesenchymal stem cells into endothelial cells under cigarette smoke extract exposure. BMC Pulm. Med. 2022, 22, 139. [Google Scholar] [CrossRef] [PubMed]
- Manderfield, L.J.; High, F.A.; Engleka, K.A.; Liu, F.; Li, L.; Rentschler, S.; Epstein, J.A. Notch Activation of Jagged1 Contributes to the Assembly of the Arterial Wall. Circulation 2012, 125, 314–323. [Google Scholar] [CrossRef] [PubMed]
- Akil, A.; Gutiérrez-García, A.K.; Guenter, R.; Rose, J.B.; Beck, A.W.; Chen, H.; Ren, B. Notch Signaling in Vascular Endothelial Cells, Angiogenesis, and Tumor Progression: An Update and Prospective. Front. Cell Dev. Biol. 2021, 9, 177. [Google Scholar] [CrossRef]
- Zouein, F.A.; Booz, G.W.; Altara, R. STAT3 and Endothelial Cell—Cardiomyocyte Dialog in Cardiac Remodeling. Front. Cardiovasc. Med. 2019, 6, 50. [Google Scholar] [CrossRef]
- Zheng, C.-X.; Wang, S.-M.; Bai, Y.-H.; Luo, T.-T.; Wang, J.-Q.; Dai, C.-Q.; Guo, B.-L.; Luo, S.-C.; Wang, D.-H.; Yang, Y.-L.; et al. Lentiviral Vectors and Adeno-Associated Virus Vectors: Useful Tools for Gene Transfer in Pain Research. Anat. Rec. 2018, 301, 825–836. [Google Scholar] [CrossRef] [Green Version]
- Yu, B.; Kiechl, S.; Qi, D.; Wang, X.; Song, Y.; Weger, S.; Mayr, A.; Le Bras, A.; Karamariti, E.; Zhang, Z.; et al. A Cytokine-Like Protein Dickkopf-Related Protein 3 Is Atheroprotective. Circulation 2017, 136, 1022–1036. [Google Scholar] [CrossRef] [Green Version]
- Gong, W.; Das, S.; Sierra-Pagan, J.E.; Skie, E.; Dsouza, N.; Larson, T.A.; Garry, M.G.; Luzete-Monteiro, E.; Zaret, K.S.; Garry, D.J. ETV2 functions as a pioneer factor to regulate and reprogram the endothelial lineage. Nat. Cell Biol. 2022, 24, 672–684. [Google Scholar] [CrossRef]
- Park, C.; Lee, T.-J.; Bhang, S.H.; Liu, F.; Nakamura, R.; Oladipupo, S.S.; Pitha-Rowe, I.; Capoccia, B.; Choi, H.S.; Kim, T.M.; et al. Injury-Mediated Vascular Regeneration Requires Endothelial ER71/ETV2. Arter. Thromb. Vasc. Biol. 2016, 36, 86–96. [Google Scholar] [CrossRef]
- Hayashi, M.; Pluchinotta, M.; Momiyama, A.; Tanaka, Y.; Nishikawa, S.-I.; Kataoka, H. Endothelialization and altered hematopoiesis by persistent Etv2 expression in mice. Exp. Hematol. 2012, 40, 738–750.e11. [Google Scholar] [CrossRef] [PubMed]
- Koyano-Nakagawa, N.; Garry, D.J. Etv2 as an essential regulator of mesodermal lineage development. Cardiovasc. Res. 2017, 113, 1294–1306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schachterle, W.; Badwe, C.R.; Palikuqi, B.; Kunar, B.; Ginsberg, M.; Lis, R.; Yokoyama, M.; Elemento, O.; Scandura, J.M.; Rafii, S. Sox17 drives functional engraftment of endothelium converted from non-vascular cells. Nat. Commun. 2017, 8, 13963. [Google Scholar] [CrossRef] [Green Version]
- Han, J.-K.; Chang, S.-H.; Cho, H.-J.; Choi, S.-B.; Ahn, H.-S.; Lee, J.; Jeong, H.; Youn, S.-W.; Lee, H.-J.; Kwon, Y.-W.; et al. Direct Conversion of Adult Skin Fibroblasts to Endothelial Cells by Defined Factors. Circulation 2014, 130, 1168–1178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hywood, J.D.; Sadeghipour, S.; Clayton, Z.E.; Yuan, J.; Stubbs, C.; Wong, J.W.T.; Cooke, J.P.; Patel, S. Induced endothelial cells from peripheral arterial disease patients and neonatal fibroblasts have comparable angiogenic properties. PLoS ONE 2021, 16, e0255075. [Google Scholar] [CrossRef]
- Ju, H.; Zhang, C.; Lu, W. Progress in heterologous biosynthesis of forskolin. J. Ind. Microbiol. Biotechnol. 2021, 48, 9. [Google Scholar] [CrossRef] [PubMed]
- Salehi, B.; Staniak, M.; Czopek, K.; Stępień, A.; Dua, K.; Wadhwa, R.; Chellappan, D.K.; Sytar, O.; Brestic, M.; Bhat, N.G.; et al. The Therapeutic Potential of the Labdane Diterpenoid Forskolin. Appl. Sci. 2019, 9, 4089. [Google Scholar] [CrossRef] [Green Version]
- Palikuqi, B.; Nguyen, D.-H.T.; Li, G.; Schreiner, R.; Pellegata, A.F.; Liu, Y.; Redmond, D.; Geng, F.; Lin, Y.; Gómez-Salinero, J.M.; et al. Adaptable haemodynamic endothelial cells for organogenesis and tumorigenesis. Nature 2020, 585, 426–432. [Google Scholar] [CrossRef]
- Lai, L.; Reineke, E.; Hamilton, D.; Cooke, J.P. Glycolytic Switch Is Required for Transdifferentiation to Endothelial Lineage. Circulation 2019, 139, 119–133. [Google Scholar] [CrossRef]
- Meng, S.; Zhou, G.; Gu, Q.; Chanda, P.K.; Ospino, F.; Cooke, J.P. Transdifferentiation Requires iNOS Activation. Circ. Res. 2016, 119, e129–e138. [Google Scholar] [CrossRef]
- Galagali, H.; Kim, J.K. The multifaceted roles of microRNAs in differentiation. Curr. Opin. Cell Biol. 2020, 67, 118–140. [Google Scholar] [CrossRef] [PubMed]
- Lawson, C.; Wolf, S. ICAM-1 signaling in endothelial cells. Pharmacol. Rep. 2009, 61, 22–32. [Google Scholar] [CrossRef]
- Kong, D.-H.; Kim, Y.K.; Kim, M.R.; Jang, J.H.; Lee, S. Emerging Roles of Vascular Cell Adhesion Molecule-1 (VCAM-1) in Immunological Disorders and Cancer. Int. J. Mol. Sci. 2018, 19, 1057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fontaine, M.; Herkenne, S.; Ek, O.; Paquot, A.; Boeckx, A.; Paques, C.; Nivelles, O.; Thiry, M.; Struman, I. Extracellular Vesicles Mediate Communication between Endothelial and Vascular Smooth Muscle Cells. Int. J. Mol. Sci. 2021, 23, 331. [Google Scholar] [CrossRef] [PubMed]
- Karamariti, E.; Margariti, A.; Winkler, B.; Wang, X.; Hong, X.; Baban, D.; Ragoussis, J.; Huang, Y.; Han, J.-D.J.; Wong, M.M.; et al. Smooth Muscle Cells Differentiated From Reprogrammed Embryonic Lung Fibroblasts Through DKK3 Signaling Are Potent for Tissue Engineering of Vascular Grafts. Circ. Res. 2013, 112, 1433–1443. [Google Scholar] [CrossRef] [Green Version]
- Rensen, S.S.; Doevendans, P.A.; van Eys, G.J. Regulation and characteristics of vascular smooth muscle cell phenotypic diversity. Neth. Heart J. 2007, 15, 100–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karamariti, E.; Zhai, C.; Yu, B.; Qiao, L.; Wang, Z.; Potter, C.M.; Wong, M.M.; Simpson, R.M.; Zhang, Z.; Wang, X.; et al. DKK3 (Dickkopf 3) Alters Atherosclerotic Plaque Phenotype Involving Vascular Progenitor and Fibroblast Differentiation Into Smooth Muscle Cells. Arter. Thromb. Vasc. Biol. 2018, 38, 425–437. [Google Scholar] [CrossRef] [Green Version]
- Hirai, H.; Yang, B.; Garcia-Barrio, M.T.; Rom, O.; Ma, P.X.; Zhang, J.; Chen, Y.E. Direct Reprogramming of Fibroblasts Into Smooth Muscle-Like Cells With Defined Transcription Factors—Brief Report. Arter. Thromb. Vasc. Biol. 2018, 38, 2191–2197. [Google Scholar] [CrossRef] [Green Version]
- Turner, E.C.; Huang, C.-L.; Govindarajan, K.; Caplice, N.M. Identification of a Klf4-dependent upstream repressor region mediating transcriptional regulation of the myocardin gene in human smooth muscle cells. Biochim. et Biophys. Acta Gene Regul. Mech. 2013, 1829, 1191–1201. [Google Scholar] [CrossRef]
- Zhou, B.; Zeng, S.; Li, N.; Yu, L.; Yang, G.; Yang, Y.; Zhang, X.; Fang, M.; Xia, J.; Xu, Y. Angiogenic Factor With G Patch and FHA Domains 1 Is a Novel Regulator of Vascular Injury. Arter. Thromb. Vasc. Biol. 2017, 37, 675–684. [Google Scholar] [CrossRef]
- Psaltis, P.J.; Simari, R.D. Vascular Wall Progenitor Cells in Health and Disease. Circ. Res. 2015, 116, 1392–1412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Jambusaria, A.; Hong, Z.; Marsboom, G.; Toth, P.T.; Herbert, B.-S.; Malik, A.B.; Rehman, J. SOX17 Regulates Conversion of Human Fibroblasts Into Endothelial Cells and Erythroblasts by Dedifferentiation Into CD34 + Progenitor Cells. Circulation 2017, 135, 2505–2523. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Cao, N.; Huang, Y.; Spencer, C.I.; Fu, J.-D.; Yu, C.; Liu, K.; Nie, B.; Xu, T.; Li, K.; et al. Expandable Cardiovascular Progenitor Cells Reprogrammed from Fibroblasts. Cell Stem Cell 2016, 18, 368–381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Pham, P.; Vu, N.B.; Nguyen, H.T.; Huynh, O.T.; Truong, M.T.-H. Significant improvement of direct reprogramming efficacy of fibroblasts into progenitor endothelial cells by ETV2 and hypoxia. Stem Cell Res. Ther. 2016, 7, 104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.Y.; Lee, H.; Kwon, Y.W.; Park, M.R.; Kim, J.H.; Kim, J.B. Etv2- and Fli1-Induced Vascular Progenitor Cells Enhance Functional Recovery in Ischemic Vascular Disease Model—Brief Report. Arter. Thromb. Vasc. Biol. 2020, 40, e105–e113. [Google Scholar] [CrossRef]
- Sidney, L.E.; Branch, M.J.; Dunphy, S.E.; Dua, H.S.; Hopkinson, A. Concise Review: Evidence for CD34 as a Common Marker for Diverse Progenitors. Stem Cells 2014, 32, 1380–1389. [Google Scholar] [CrossRef] [Green Version]
- Van Pham, P.; Vu, N.B.; Dao, T.T.-T.; Le, H.T.-N.; Phi, L.T.; Phan, N.K. Production of endothelial progenitor cells from skin fibroblasts by direct reprogramming for clinical usages. Vitr. Cell. Dev. Biol. Anim. 2017, 53, 207–216. [Google Scholar] [CrossRef]
- Jochems, C.E.A.; van der Valk, J.B.; Stafleu, F.R.; Baumans, V. The Use of Fetal Bovine Serum: Ethical or Scientific Problem? Altern. Lab. Anim. 2002, 30, 219–227. [Google Scholar] [CrossRef]
- Blum, K.M.; Zbinden, J.C.; Ramachandra, A.B.; Lindsey, S.E.; Szafron, J.M.; Reinhardt, J.W.; Heitkemper, M.; Best, C.A.; Mirhaidari, G.J.M.; Chang, Y.-C.; et al. Tissue engineered vascular grafts transform into autologous neovessels capable of native function and growth. Commun. Med. 2022, 2, 3. [Google Scholar] [CrossRef]
- Mallis, P.; Kostakis, A.; Stavropoulos-Giokas, C.; Michalopoulos, E. Future Perspectives in Small-Diameter Vascular Graft Engineering. Bioengineering 2020, 7, 160. [Google Scholar] [CrossRef]
- Matsuzaki, Y.; John, K.; Shoji, T.; Shinoka, T. The Evolution of Tissue Engineered Vascular Graft Technologies: From Preclinical Trials to Advancing Patient Care. Appl. Sci. 2019, 9, 1274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Generali, M.; Casanova, E.A.; Kehl, D.; Wanner, D.; Hoerstrup, S.P.; Cinelli, P.; Weber, B. Autologous endothelialized small-caliber vascular grafts engineered from blood-derived induced pluripotent stem cells. Acta Biomater. 2019, 97, 333–343. [Google Scholar] [CrossRef] [PubMed]
- Saito, J.; Kaneko, M.; Ishikawa, Y.; Yokoyama, U. Challenges and Possibilities of Cell-Based Tissue-Engineered Vascular Grafts. Cyborg Bionic Syst. 2021, 2021, 1532103. [Google Scholar] [CrossRef] [PubMed]
- Hu, K.; Li, Y.; Ke, Z.; Yang, H.; Lu, C.; Li, Y.; Guo, Y.; Wang, W. History, progress and future challenges of artificial blood vessels: A narrative review. Biomater. Transl. 2022, 3, 81. [Google Scholar] [CrossRef]
- Moore, M.J.; Tan, R.P.; Yang, N.; Rnjak-Kovacina, J.; Wise, S.G. Bioengineering artificial blood vessels from natural materials. Trends Biotechnol. 2022, 40, 693–707. [Google Scholar] [CrossRef]
- Obiweluozor, F.O.; Emechebe, G.A.; Kim, D.-W.; Cho, H.-J.; Park, C.H.; Kim, C.S.; Jeong, I.S. Considerations in the Development of Small-Diameter Vascular Graft as an Alternative for Bypass and Reconstructive Surgeries: A Review. Cardiovasc. Eng. Technol. 2020, 11, 495–521. [Google Scholar] [CrossRef]
- Liao, J.; Xu, B.; Zhang, R.; Fan, Y.; Xie, H.; Li, X. Applications of decellularized materials in tissue engineering: Advantages, drawbacks and current improvements, and future perspectives. J. Mater. Chem. B 2020, 8, 10023–10049. [Google Scholar] [CrossRef]
- Jin, Y.; Cho, S.-W. Bioengineering platforms for cell therapeutics derived from pluripotent and direct reprogramming. APL Bioeng. 2021, 5, 031501. [Google Scholar] [CrossRef]
- Jiang, Y.; Li, R.; Han, C.; Huang, L. Extracellular matrix grafts: From preparation to application (Review). Int. J. Mol. Med. 2021, 47, 463–474. [Google Scholar] [CrossRef]
- Wang, X.; Chan, V.; Corridon, P.R. Decellularized blood vessel development: Current state-of-the-art and future directions. Front. Bioeng. Biotechnol. 2022, 10, 951644. [Google Scholar] [CrossRef]
- Khanna, A.; Zamani, M.; Huang, N.F. Extracellular Matrix-Based Biomaterials for Cardiovascular Tissue Engineering. J. Cardiovasc. Dev. Dis. 2021, 8, 137. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.; Atchison, L.; Chen, Z.; Chakraborty, S.; Jung, Y.; Truskey, G.A.; Christoforou, N.; Leong, K.W. Transdifferentiation of human endothelial progenitors into smooth muscle cells. Biomaterials 2016, 85, 180–194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, Y.; Lee, J.S.; Kim, J.; Min, S.; Wi, S.; Yu, J.H.; Chang, G.-E.; Cho, A.-N.; Choi, Y.; Ahn, D.-H.; et al. Three-dimensional brain-like microenvironments facilitate the direct reprogramming of fibroblasts into therapeutic neurons. Nat. Biomed. Eng. 2018, 2, 522–539. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Kim, J.; Lee, J.S.; Min, S.; Kim, S.; Ahn, D.-H.; Kim, Y.-G.; Cho, S.-W. Vascularized Liver Organoids Generated Using Induced Hepatic Tissue and Dynamic Liver-Specific Microenvironment as a Drug Testing Platform. Adv. Funct. Mater. 2018, 28, 1801954. [Google Scholar] [CrossRef]
- Chen, S.-G.; Ugwu, F.; Li, W.-C.; Caplice, N.M.; Petcu, E.B.; Yip, S.P.; Huang, C.-L. Vascular Tissue Engineering: Advanced Techniques and Gene Editing in Stem Cells for Graft Generation. Tissue Eng. Part B Rev. 2021, 27, 14–28. [Google Scholar] [CrossRef]
- Liguori, G.; Sinkunas, V.; Liguori, T.; Moretto, E.; Sharma, P.; Harmsen, M.; Moreira, L. Decellularized Arterial Extracellular Matrix-Based Hydrogel Supports 3D Bioprinting of the Media Layer of Small-Caliber Blood Vessels|Circulation. Circ. Cell Tissue Eng. 2019, 140, A14119. [Google Scholar]
- Ho, L.; Hsu, S.-H. Cell reprogramming by 3D bioprinting of human fibroblasts in polyurethane hydrogel for fabrication of neural-like constructs. Acta Biomater. 2018, 70, 57–70. [Google Scholar] [CrossRef]
- Lui, C.; Chin, A.F.; Park, S.; Yeung, E.; Kwon, C.; Tomaselli, G.; Chen, Y.; Hibino, N. Mechanical stimulation enhances development of scaffold-free, 3D-printed, engineered heart tissue grafts. J. Tissue Eng. Regen. Med. 2021, 15, 503–512. [Google Scholar] [CrossRef]
- Sato, Y.; Yamamoto, K.; Horiguchi, S.; Tahara, Y.; Nakai, K.; Kotani, S.-I.; Oseko, F.; Pezzotti, G.; Yamamoto, T.; Kishida, T.; et al. Nanogel tectonic porous 3D scaffold for direct reprogramming fibroblasts into osteoblasts and bone regeneration. Sci. Rep. 2018, 8, 15824. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Hu, J.; Jiao, J.; Liu, Z.; Zhou, Z.; Zhao, C.; Chang, L.-J.; Chen, Y.E.; Ma, P.X.; Yang, B. Engineering vascular tissue with functional smooth muscle cells derived from human iPS cells and nanofibrous scaffolds. Biomaterials 2014, 35, 8960–8969. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carlson, A.L.; Bennett, N.; Francis, N.; Halikere, A.; Clarke, S.; Moore, J.C.; Hart, R.; Paradiso, K.; Wernig, M.; Kohn, J.; et al. Generation and transplantation of reprogrammed human neurons in the brain using 3D microtopographic scaffolds. Nat. Commun. 2016, 7, 10862. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Xue, F.; Zhang, H.; Sanyour, H.J.; Rickel, A.P.; Uttecht, A.; Fanta, B.; Hu, J.; Hong, Z. Fabrication and Characterization of Pectin Hydrogel Nanofiber Scaffolds for Differentiation of Mesenchymal Stem Cells into Vascular Cells. ACS Biomater. Sci. Eng. 2019, 5, 6511–6519. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Huang, Y.; Pang, M.; Yang, Y.; Linshan, L.; Liu, L.; Shu, T.; Zhou, W.; Wang, X.; Rong, L.; et al. Tissue-Engineered Regeneration of Completely Transected Spinal Cord Using Induced Neural Stem Cells and Gelatin-Electrospun Poly (Lactide-Co-Glycolide)/Polyethylene Glycol Scaffolds. PLoS ONE 2015, 10, e0117709. [Google Scholar] [CrossRef] [Green Version]
- Haddad, T.; Noel, S.; Liberelle, B.; El Ayoubi, R.; Ajji, A.; De Crescenzo, G. Fabrication and surface modification of poly lactic acid (PLA) scaffolds with epidermal growth factor for neural tissue engineering. Biomatter 2016, 6, e1231276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lo, B.; Parham, L. Ethical Issues in Stem Cell Research. Endocr. Rev. 2009, 30, 204–213. [Google Scholar] [CrossRef] [PubMed]
- Byrnes, W.M.; Center, T.N.C.B. Direct Reprogramming and Ethics in Stem Cell Research. Natl. Cathol. Bioeth. Q. 2008, 8, 277–290. [Google Scholar] [CrossRef]
- Dotson, G.A.; Ryan, C.W.; Chen, C.; Muir, L.; Rajapakse, I. Cellular reprogramming: Mathematics meets medicine. WIREs Mech. Dis. 2020, 13, e1515. [Google Scholar] [CrossRef]
- Tran, A.; Yang, P.; Yang, J.Y.H.; Ormerod, J. Computational approaches for direct cell reprogramming: From the bulk omics era to the single cell era. Brief. Funct. Genom. 2022, 21, 270–279. [Google Scholar] [CrossRef]
- Eguchi, R.; Hamano, M.; Iwata, M.; Nakamura, T.; Oki, S.; Yamanishi, Y. TRANSDIRE: Data-driven direct reprogramming by a pioneer factor-guided trans-omics approach. Bioinformatics 2022, 38, 2839–2846. [Google Scholar] [CrossRef]
- Rackham, O.; The FANTOM Consortium; Firas, J.; Fang, H.; Oates, M.E.; Holmes, M.; Knaupp, A.; Suzuki, H.; Nefzger, C.; Daub, C.O.; et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016, 48, 331–335. [Google Scholar] [CrossRef] [Green Version]
- Ronquist, S.; Patterson, G.; Muir, L.A.; Lindsly, S.; Chen, H.; Brown, M.; Wicha, M.S.; Bloch, A.; Brockett, R.; Rajapakse, I. Algorithm for cellular reprogramming. Proc. Natl. Acad. Sci. USA 2017, 114, 11832–11837. [Google Scholar] [CrossRef] [PubMed]
- Gam, R.; Sung, M.; Pandurangan, A.P. Experimental and Computational Approaches to Direct Cell Reprogramming: Recent Advancement and Future Challenges. Cells 2019, 8, 1189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guerrero-Ramirez, G.; Valdez-Cordoba, C.; Islas-Cisneros, J.; Trevino, V. Computational approaches for predicting key transcription factors in targeted cell reprogramming (Review). Mol. Med. Rep. 2018, 18, 1225–1237. [Google Scholar] [CrossRef] [PubMed]
- Majesky, M.W. Developmental Basis of Vascular Smooth Muscle Diversity. Arter. Thromb. Vasc. Biol. 2007, 27, 1248–1258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, G.; Jacquet, L.; Karamariti, E.; Xu, Q. Origin and differentiation of vascular smooth muscle cells. J. Physiol. 2015, 593, 3013–3030. [Google Scholar] [CrossRef] [Green Version]
- Shen, M.; Quertermous, T.; Fischbein, M.P.; Wu, J.C. Generation of Vascular Smooth Muscle Cells From Induced Pluripotent Stem Cells. Circ. Res. 2021, 128, 670–686. [Google Scholar] [CrossRef]
- Topouzis, S.; Majesky, M.W. Smooth Muscle Lineage Diversity in the Chick Embryo. Two types of aortic smooth muscle cell differ in growth and receptor-mediated transcriptional responses to transforming growth factor-beta. Dev. Biol. 1996, 178, 430–445. [Google Scholar] [CrossRef]
- Wong, A.P.; Nili, N.; Strauss, B.H. In vitro differences between venous and arterial-derived smooth muscle cells: Potential modulatory role of decorin. Cardiovasc. Res. 2005, 65, 702–710. [Google Scholar] [CrossRef]
- Cuenca, M.V.; Cochrane, A.; Hil, F.E.V.D.; de Vries, A.A.; Oberstein, S.A.L.; Mummery, C.L.; Orlova, V.V. Engineered 3D vessel-on-chip using hiPSC-derived endothelial- and vascular smooth muscle cells. Stem Cell Rep. 2021, 16, 2159–2168. [Google Scholar] [CrossRef]
- Yao, M.; Ren, T.; Pan, Y.; Xue, X.; Li, R.; Zhang, L.; Li, Y.; Huang, K. A New Generation of Lineage Tracing Dynamically Records Cell Fate Choices. Int. J. Mol. Sci. 2022, 23, 5021. [Google Scholar] [CrossRef]
- Zhang, Y.; Zeng, F.; Han, X.; Weng, J.; Gao, Y. Lineage tracing: Technology tool for exploring the development, regeneration, and disease of the digestive system. Stem Cell Res. Ther. 2020, 11, 438. [Google Scholar] [CrossRef] [PubMed]
- Alemany, A.; Florescu, M.; Baron, C.S.; Peterson-Maduro, J.; van Oudenaarden, A. Whole-organism clone tracing using single-cell sequencing. Nature 2018, 556, 108–112. [Google Scholar] [CrossRef] [PubMed]
- Schlimgen, R.; Howard, J.; Wooley, D.; Thompson, M.; Baden, L.R.; Yang, O.O.; Christiani, D.C.; Mostoslavsky, G.; Diamond, D.V.; Duane, E.G.; et al. Risks Associated With Lentiviral Vector Exposures and Prevention Strategies. J. Occup. Environ. Med. 2016, 58, 1159–1166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Connolly, J.B. Lentiviruses in gene therapy clinical research. Gene Ther. 2002, 9, 1730–1734. [Google Scholar] [CrossRef] [Green Version]
- Rubio, A.; Luoni, M.; Giannelli, S.G.; Radice, I.; Iannielli, A.; Cancellieri, C.; Di Berardino, C.; Regalia, G.; Lazzari, G.; Menegon, A.; et al. Rapid and efficient CRISPR/Cas9 gene inactivation in human neurons during human pluripotent stem cell differentiation and direct reprogramming. Sci. Rep. 2016, 6, 37540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sokka, J.; Yoshihara, M.; Kvist, J.; Laiho, L.; Warren, A.; Stadelmann, C.; Jouhilahti, E.-M.; Kilpinen, H.; Balboa, D.; Katayama, S.; et al. CRISPR activation enables high-fidelity reprogramming into human pluripotent stem cells. Stem Cell Rep. 2022, 17, 413–426. [Google Scholar] [CrossRef] [PubMed]
- Weltner, J.; Balboa, D.; Katayama, S.; Bespalov, M.; Krjutškov, K.; Jouhilahti, E.-M.; Trokovic, R.; Kere, J.; Otonkoski, T. Human pluripotent reprogramming with CRISPR activators. Nat. Commun. 2018, 9, 2643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, L.; Liang, J.; Huang, W.; Ma, J.; Park, K.H.; Wu, Z.; Chen, P.; Zhu, H.; Ma, J.-J.; Cai, W.; et al. CRISPR activation of endogenous genes reprograms fibroblasts into cardiovascular progenitor cells for myocardial infarction therapy. Mol. Ther. 2022, 30, 54–74. [Google Scholar] [CrossRef]
- Deng, J.; Luo, K.; Xu, P.; Jiang, Q.; Wang, Y.; Yao, Y.; Chen, X.; Cheng, F.; Xie, D.; Deng, H. High-efficiency c-Myc-mediated induction of functional hepatoblasts from the human umbilical cord mesenchymal stem cells. Stem Cell Res. Ther. 2021, 12, 375. [Google Scholar] [CrossRef]
- Wernig, M.; Meissner, A.; Cassady, J.P.; Jaenisch, R. c-Myc Is Dispensable for Direct Reprogramming of Mouse Fibroblasts. Cell Stem Cell 2008, 2, 10–12. [Google Scholar] [CrossRef] [Green Version]
- Bersini, S.; Schulte, R.; Huang, L.; Tsai, H.; Hetzer, M.W. Direct reprogramming of human smooth muscle and vascular endothelial cells reveals defects associated with aging and Hutchinson-Gilford progeria syndrome. Elife 2020, 9, e54383. [Google Scholar] [CrossRef] [PubMed]
- Maffioletti, S.M.; Sarcar, S.; Henderson, A.B.; Mannhardt, I.; Pinton, L.; Moyle, L.A.; Steele-Stallard, H.; Cappellari, O.; Wells, K.E.; Ferrari, G.; et al. Three-Dimensional Human iPSC-Derived Artificial Skeletal Muscles Model Muscular Dystrophies and Enable Multilineage Tissue Engineering. Cell Rep. 2018, 23, 899–908. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Dal-Pra, S.; Mirotsou, M.; Jayawardena, T.M.; Hodgkinson, C.P.; Bursac, N.; Dzau, V.J. Tissue-engineered 3-dimensional (3D) microenvironment enhances the direct reprogramming of fibroblasts into cardiomyocytes by microRNAs. Sci. Rep. 2016, 6, 38815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, A.W.; Hoyne, J.D.; Nguyen, P.K.; McCreedy, D.A.; Aly, H.; Efimov, I.R.; Rentschler, S.; Elbert, D.L. Direct reprogramming of mouse fibroblasts to cardiomyocyte-like cells using Yamanaka factors on engineered poly(ethylene glycol) (PEG) hydrogels. Biomaterials 2013, 34, 6559–6571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paoletti, C.; Divieto, C.; Chiono, V. Impact of Biomaterials on Differentiation and Reprogramming Approaches for the Generation of Functional Cardiomyocytes. Cells 2018, 7, 114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moses, S.R.; Adorno, J.J.; Palmer, A.F.; Song, J.W. Vessel-on-a-chip models for studying microvascular physiology, transport, and function in vitro. Am. J. Physiol. Physiol. 2021, 320, C92–C105. [Google Scholar] [CrossRef]
- Kim, S.; Kim, W.; Lim, S.; Jeon, J.S. Vasculature-On-A-Chip for In Vitro Disease Models. Bioengineering 2017, 4, 8. [Google Scholar] [CrossRef] [Green Version]





| Reference | Source Cells | Transcription Factors | Culture Medium | Functional Outcome | Therapeutic Potential | Signalling Pathway | Limitations |
|---|---|---|---|---|---|---|---|
| Margariti et al., 2012 [10] | Human embryonic lung fibroblasts (HELF) | OCT4, SOX2, KLF4, C-MYC | EGM2 | iECs were stable and formed patent vessels when constituted onto a decellularised vessel scaffold | Hindlimb ischaemia: increased capillary number and blood perfusion | SETSIP activation which promotes EC-specific gene expression | Embryonic cell source is ethically controversial |
| Li et al., 2013 [11] | Human neonatal fibroblasts | OCT4, KLF4 | Differentiation medium II (50 ng/mL VEGF, 20 ng/mL bFGF, 0.1 mM 8-Br-CAMP) | Addition of 8-Br-cAMP increased transdifferentiation of fibroblasts into iECs | Murine hindlimb ischaemic model observed increased capillary number and blood perfusion | Not assessed | Conversion efficacy was low compared to studies using all 4 OSKM factors prior to sorting methods |
| Hong et al., 2017 [12] | Human umbilical artery smooth muscle cells | OCT4, SOX2, KLF4, C-MYC | EGM-2 (CC-3162) plus 25 ng/mL VEGF | SMCs are capable of trans-differentiation to iECs | Murine hindlimb ischaemic model observed increased capillary number and blood perfusion | OSKM upregulates VE-cadherin, HES4 and JAG1 which increases EC-specific gene expression | Lentiviral vectors possess safety risks. Viability of plasmid delivery confirmed but not explored |
| Chen et al., 2019 [13] | HELF | DKK3 | EGM-2 plus EGM BulletKit (CC-3124) plus 10 ng/mL VEGF | iECs formed a patent monolayer in an ex vivo vascular graft | Formation of microvascular structures in vivo | Increases MET and VEGFR2, decreases miR-125a-5p and promotes Stat3 | Embryonic cell source is ethically controversial |
| Ginsberg et al., 2012 [14] | Human amniotic fluid-derived cells | ETV2, FLI1, ERG1, TGFβ inhibition | EM (Medium 199, 15% FBS, 20 μg/mL endothelial cell supplement, 1X Pen/Strep, 20 units/mL Heparin) | iECs were generic and may hold potential for further subtype specification | In a regenerating mouse liver model, engraftment of iECs resulted in patent capillaries | Not assessed | Unsuccessfully with human postnatal cells. Use of amniotic cells is ethically controversial |
| Wong and Cooke, 2016 [15] | Human neonatal foreskin fibroblasts | ETV2, FLI1, GATA2, KLF4 | EGM-2 plus 10 µmol/L SB341542 | iECs uptake Ac-LDL and formed capillary-like networks | Not assessed | Not assessed | Therapeutic potential unknown |
| Kim et al., 2020 [16] | Human dermal fibroblasts | ETV2 | EGM-2 plus doxycycline | iECs formed stable endothelial layers when seeded on a decellularised liver scaffold | Hindlimb ischaemia: improved angiogenic capabilities and blood perfusion | cAMP/EPAC/RAP1 | iECs were not easily expandable |
| Morita et al., 2015 [17] | Human dermal fibroblasts | ETV2 | EGM-2 medium (10 ng/mL recombinant human VEGF165, bFGF) | iECs displayed venous properties but adopted arteriole characteristics when combined with mural cells | Hindlimb ischaemia: improved angiogenic capabilities and blood perfusion | Modify DNA methylation states of EC genes | Extensive 50-day ETV2 exposure, lacking maturity, failed to induce NOS3 |
| Lee et al., 2017 [18] | Human postnatal dermal fibroblasts | ETV2 | EGM-2 plus DOX | Generation of early immature iECs, followed by matured iECs | Injection of early iECs into a murine hindlimb ischaemic model improved vessel generation and tissue perfusion | Not assessed | Early immature iECs failed to direct incorporation into host vasculature. Long timeline to cultivate mature iECs |
| Sayed et al., 2015 [9] | Human neonatal foreskin fibroblasts | Poly I:C (TLR3 agonist) | Maintenance medium (bFGF, VEGF, 0.1 mmol/L 8-Br-cAMP) | Innate immune activation is necessary for human fibroblasts to transdifferentiate into ECs effectively | Murine hindlimb ischaemic model observed increased expansion of host vasculature, blood perfusion and decreased tissue injury | Innate immune activation, TLR3/NF-κB/iNOS, epigenetic plasticity. Metabolic switching from oxidative phosphorylation to glycolysis | Low transdifferentiation efficacy. Therapeutic potential unknown, metabolic heterogeneity in iECs |
| McCoy et al., 2022 [19] | Human coronary artery smooth muscle cells (CASMCs) | miRNA | EGM-2 (2 µL/2 mL 8-Br-cAMP, 2 µL/mL SB 431542) | iECs exhibit high similarity to native ECs | Quicker limb reperfusion | Upregulation of NOTCH1, JAG1, and DLL4 | Other miRNA targets need to be explored further |
| Reference | Source Cell | Transcription Factors | Culture Medium | Functional Outcome | In Vivo Therapeutic Potential | Signalling Pathway | Limitations |
|---|---|---|---|---|---|---|---|
| Karamariti et al., 2013 [45] | HELF | OCT4, SOX2, KLF4, C-MYC | DM (MEM α, 10% FBS, 100 U/mL penicillin and streptomycin, 0.2 mM L-glutamine, 0.1 mM β-mercaptoethanol, 10 ng/mL PDGF-BB) | iVSMCs | Transplantation of iVSMCs-seeded decellularised vessel in mice increased survival | DKK3/Kremen1/Wnt signalling | Limited to HELF, Unknown efficacy of iVSMC generation, HELF is ethically controversial |
| Karamariti et al., 2018 [47] | HELF | DKK3 | DMEM (ATCC, 10% EmbryoMax® ES Cell Qualified FBS, 10 ng/mL LIF, 0.1 mM 2-mercaptoethanol) on a 0.04% gelatin substrate | VPCs, iVSMCs | Promotes stabilisation of atherosclerotic plaques by increasing SMCs and suppressing inflammation | DKK3/ATF6/TGFβ1 | HELF is ethically controversial. |
| Hirai et al., 2018 [48] | MEF and adult dermal fibroblasts | Myocd, GATA6, MEF2C | SMC medium (DMEM/F-12, 10% KSR, 2 ng/mL recombinant human TGF-β1, 10 ng/mL human PDGF-BB, 1% penicillin-streptomycin) | iVSMCs | Not assessed | Not assessed | Partially reprogrammed iVSMCs |
| Reference | Source Cell | Transcription Factors | Culture Medium | Functional Outcome | In Vivo Therapeutic Potential | Signalling Pathway | Limitations |
|---|---|---|---|---|---|---|---|
| Kurian et al., 2013 [8] | Human neonatal and adult fibroblasts | OCT4, SOX2, KLF4, C-MYC | MIM (DMEM:F12,15 mg mL−1 stem cell–grade BSA, 17.5 μg mL−1 human insulin, 275 μg mL−1 human holo-transferrin, 20 ng mL−1 bFGF, 50 ng mL−1 human VEGF-165 aa, 25 ng mL−1 human BMP4, 450 μM monothioglycerol, 2.25 mM L-glutamine, 2.25 mM NEAA) | CD34+ angioblast-like bipotent progenitors | Forming functional blood vessels that integrated with host vasculature | Not investigated | Heterogenous cells |
| Zhang et al., 2017a [52] | Human adult and neonatal dermal fibroblast | OCT4, SOX2, KLF4, C-MYC | DMEM/F12 (20% KSR,10 ng mL−1 bFGF, 1 mM GlutaMAX, 0.1 mM NEAA, 55 μM β-mercaptoethanol) | Induced tripotent cardiac progenitor cells (iSMCs, iECs, iCMs) | Improved cardiac function and reduced adverse cardiac remodelling | Not investigated | Teratoma risk |
| Zhang et al., 2016 [53] | MEF | OCT4, SOX2, KLF4, C-MYC | ieCPC basal medium plus Advanced DMEM/F12: Neural basal (1:1) (1X N2, 1X B27 without Vitamin A, 1X Glutamax, 1X NEAA, 0.05% BSA, 0.1 mM β-ME) plus BACS, (5 ng/mL BMP4, 10 ng/mL Activin A, 3 μM CHIR99021, 2 μM SU5402) | BACS as a reliable prerequisite for the effective creation and ongoing renewal of ieCPCs | Directly produce CMs, ECs, and SMCs when exposed to the infarcted heart environment in vivo | Not investigated | Translational applicability of these cells |
| Pham et al., 2016 [54] | Human dermal fibroblasts | ETV2 | Medium 200 (5% PRP, 5 ng/mL recombinant EGF, 1 ng/mL recombinant VEGF, 20 ng/mL insulin-like growth factor, 1 μg/mL ascorbic acid, 0.2 μg/ mL hydrocortisone, 22.5 μg/mL heparin, 1% antibiotic-antimycotic) | Unipotent iEPCs | Improve hindlimb ischemia | Not investigated | Venous not arterial ECs |
| Park et al., 2020 [55] | Mouse fibroblasts | ETV2, Fli1 | VPC medium (10% FBS, 2 mmol/L L-glutamine, β-mercaptoethanol, penicillin/streptomycin, 10 ng/mL VEGF) | Self-renewal and biopotency iVPCs | Enhanced blood flow without tumour formation | Not investigated | Contamination of residual undifferentiated PSC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sellahewa, S.G.; Li, J.Y.; Xiao, Q. Updated Perspectives on Direct Vascular Cellular Reprogramming and Their Potential Applications in Tissue Engineered Vascular Grafts. J. Funct. Biomater. 2023, 14, 21. https://doi.org/10.3390/jfb14010021
Sellahewa SG, Li JY, Xiao Q. Updated Perspectives on Direct Vascular Cellular Reprogramming and Their Potential Applications in Tissue Engineered Vascular Grafts. Journal of Functional Biomaterials. 2023; 14(1):21. https://doi.org/10.3390/jfb14010021
Chicago/Turabian StyleSellahewa, Saneth Gavishka, Jojo Yijiao Li, and Qingzhong Xiao. 2023. "Updated Perspectives on Direct Vascular Cellular Reprogramming and Their Potential Applications in Tissue Engineered Vascular Grafts" Journal of Functional Biomaterials 14, no. 1: 21. https://doi.org/10.3390/jfb14010021