Oxylipins and Reactive Carbonyls as Regulators of the Plant Redox and Reactive Oxygen Species Network under Stress
Abstract
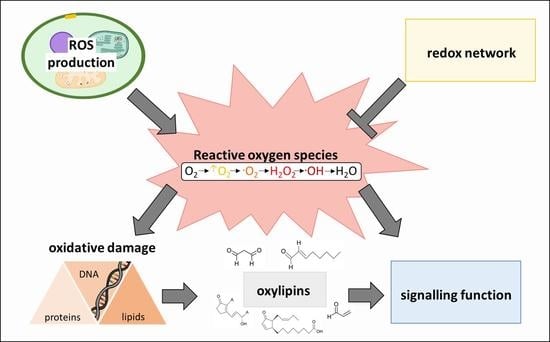
:1. Introduction
2. The Plant ROS and Redox Network
2.1. Synthesis of ROS in Higher Plants
2.2. ROS Homeostasis as Key Mechanism to Avoid Oxidative Stress
2.2.1. Structure of the Redox-Regulatory Network
2.2.2. Exemplary Function of the Redox Network: The Water-Water-Cycle
2.3. Decomposition of ROS
3. Oxylipins
3.1. Non-Enzymatic and Enzymatic Lipid Peroxidation Yields Highly Diverse Oxylipins
3.1.1. Phytoprostanes Are Evolutionary Ancient Oxylipins
3.1.2. 12-OPDA and OPDAylation as Potent PTM
3.1.3. Oxylipin Aldehydes
3.2. Oxylipin Signature
4. Influence of Oxylipins on the Redox-Regulatory Network
4.1. Modulation of Thiol-Sensitive Proteins
4.2. Interaction with Non-Protein Thiols
4.3. Contribution of Oxylipins to Environmental Acclimatization
4.3.1. Thermotolerance
4.3.2. Pathogen Infection and Induced Systemic Resistance (ISR)
4.3.3. Flooding
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Holland, H.D. The oxygenation of the atmosphere and oceans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2006, 361, 903–915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vogelsang, L.; Dietz, K.-J. Plant thiol peroxidases as redox sensors and signal transducers in abiotic stress acclimation. Free Radic. Biol. Med. 2022, 193, 764–778. [Google Scholar] [CrossRef] [PubMed]
- Turkan, I. Emerging roles for ROS and RNS—Versatile molecules in plants. J. Exp. Bot. 2017, 68, 4413–4416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mittler, R.; Zandalinas, S.I.; Fichman, Y.; van Breusegem, F. Reactive oxygen species signalling in plant stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 663–679. [Google Scholar] [CrossRef]
- Juan, C.A.; La Pérez de Lastra, J.M.; Plou, F.J.; Pérez-Lebeña, E. The Chemistry of Reactive Oxygen Species (ROS) Revisited: Outlining Their Role in Biological Macromolecules (DNA, Lipids and Proteins) and Induced Pathologies. Int. J. Mol. Sci. 2021, 22, 4642. [Google Scholar] [CrossRef]
- Khan, M.; Ali, S.; Al Azzawi, T.N.I.; Saqib, S.; Ullah, F.; Ayaz, A.; Zaman, W. The Key Roles of ROS and RNS as a Signaling Molecule in Plant–Microbe Interactions. Antioxidants 2023, 12, 268. [Google Scholar] [CrossRef]
- Tripathy, B.C.; Oelmüller, R. Reactive oxygen species generation and signaling in plants. Plant Signal. Behav. 2012, 7, 1621–1633. [Google Scholar] [CrossRef] [Green Version]
- Dumanović, J.; Nepovimova, E.; Natić, M.; Kuča, K.; Jaćević, V. The Significance of Reactive Oxygen Species and Antioxidant Defense System in Plants: A Concise Overview. Front. Plant Sci. 2020, 11, 552969. [Google Scholar] [CrossRef]
- Asada, K. Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol. 2006, 141, 391–396. [Google Scholar] [CrossRef] [Green Version]
- Farooq, M.A.; Niazi, A.K.; Akhtar, J.; Saifullah; Farooq, M.; Souri, Z.; Karimi, N.; Rengel, Z. Acquiring control: The evolution of ROS-Induced oxidative stress and redox signaling pathways in plant stress responses. Plant Physiol. Biochem. 2019, 141, 353–369. [Google Scholar] [CrossRef]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef]
- Bhattacharjee, S. Reactive oxygen species and oxidative burst: Roles in stress, senescence and signal transduction in plants. Curr. Sci. 2005, 89, 1113–1121. [Google Scholar]
- Gerken, M.; Kakorin, S.; Chibani, K.; Dietz, K.-J. Computational simulation of the reactive oxygen species and redox network in the regulation of chloroplast metabolism. PLoS Comput. Biol. 2020, 16, e1007102. [Google Scholar] [CrossRef] [Green Version]
- Sies, H. Oxidative eustress: On constant alert for redox homeostasis. Redox Biol. 2021, 41, 101867. [Google Scholar] [CrossRef]
- Khorobrykh, S.; Havurinne, V.; Mattila, H.; Tyystjärvi, E. Oxygen and ROS in Photosynthesis. Plants 2020, 9, 91. [Google Scholar] [CrossRef] [Green Version]
- Mittler, R. ROS Are Good. Trends Plant Sci. 2017, 22, 11–19. [Google Scholar] [CrossRef] [Green Version]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; van Breusegem, F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef]
- Fridovich, I. Biological effects of the superoxide radical. Arch. Biochem. Biophys. 1986, 247, 1–11. [Google Scholar] [CrossRef]
- Noctor, G.; Veljovic-Jovanovic, S.; Driscoll, S.; Novitskaya, L.; Foyer, C.H. Drought and oxidative load in the leaves of C3 plants: A predominant role for photorespiration? Ann. Bot. 2002, 89, 841–850. [Google Scholar] [CrossRef]
- Corpas, F.J.; Barroso, J.B. Peroxisomal plant metabolism—An update on nitric oxide, Ca2+ and the NADPH recycling network. J. Cell Sci. 2018, 131. [Google Scholar] [CrossRef] [Green Version]
- Corpas, F.J.; González-Gordo, S.; Palma, J.M. Plant Peroxisomes: A Factory of Reactive Species. Front. Plant Sci. 2020, 11, 853. [Google Scholar] [CrossRef] [PubMed]
- Corpas, F.J.; Barroso, J.B.; Palma, J.M.; Rodriguez-Ruiz, M. Plant peroxisomes: A nitro-oxidative cocktail. Redox Biol. 2017, 11, 535–542. [Google Scholar] [CrossRef] [PubMed]
- Corpas, F.J.; Del Río, L.A.; Palma, J.M. Plant peroxisomes at the crossroad of NO and H2O2 metabolism. J. Integr. Plant Biol. 2019, 61, 803–816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mano, J.; Biswas, M.S.; Sugimoto, K. Reactive Carbonyl Species: A Missing Link in ROS Signaling. Plants 2019, 8, 391. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, K.; Yokochi, Y.; Tanaka, K.; Hisabori, T. The ferredoxin/thioredoxin pathway constitutes an indispensable redox-signaling cascade for light-dependent reduction of chloroplast stromal proteins. J. Biol. Chem. 2022, 298, 102650. [Google Scholar] [CrossRef]
- Liebthal, M.; Maynard, D.; Dietz, K.-J. Peroxiredoxins and Redox Signaling in Plants. Antioxid. Redox Signal. 2018, 28, 609–624. [Google Scholar] [CrossRef] [Green Version]
- Maynard, D.; Gröger, H.; Dierks, T.; Dietz, K.-J. The function of the oxylipin 12-oxophytodienoic acid in cell signaling, stress acclimation, and development. J. Exp. Bot. 2018, 69, 5341–5354. [Google Scholar] [CrossRef]
- Park, S.-W.; Li, W.; Viehhauser, A.; He, B.; Kim, S.; Nilsson, A.K.; Andersson, M.X.; Kittle, J.D.; Ambavaram, M.M.R.; Luan, S.; et al. Cyclophilin 20-3 relays a 12-oxo-phytodienoic acid signal during stress responsive regulation of cellular redox homeostasis. Proc. Natl. Acad. Sci. USA 2013, 110, 9559–9564. [Google Scholar] [CrossRef] [Green Version]
- Laxa, M.; König, J.; Dietz, K.-J.; Kandlbinder, A. Role of the cysteine residues in Arabidopsis thaliana cyclophilin CYP20-3 in peptidyl-prolyl cis-trans isomerase and redox-related functions. Biochem. J. 2007, 401, 287–297. [Google Scholar] [CrossRef] [Green Version]
- Feldman-Salit, A.; Wirtz, M.; Hell, R.; Wade, R.C. A mechanistic model of the cysteine synthase complex. J. Mol. Biol. 2009, 386, 37–59. [Google Scholar] [CrossRef]
- Asada, K.; Kiso, K.; Yoshikawa, K. Univalent Reduction of Molecular Oxygen by Spinach Chloroplasts on Illumination. J. Biol. Chem. 1974, 249, 2175–2181. [Google Scholar] [CrossRef]
- Mehler, A.H. Studies on reactions of illuminated chloroplasts. I. Mechanism of the reduction of oxygen and other Hill reagents. Arch. Biochem. Biophys. 1951, 33, 65–77. [Google Scholar] [CrossRef]
- Mano, J.; Endo, T.; Miyake, C. How do photosynthetic organisms manage light stress? A tribute to the late Professor Kozi Asada. Plant Cell Physiol. 2016, 57, 1351–1353. [Google Scholar] [CrossRef] [Green Version]
- Ishikawa, T.; Maruta, T.; Ogawa, T.; Yoshimura, K.; Shigeoka, S. Redox Balance in Chloroplasts as a Modulator of Environmental Stress Responses: The Role of Ascorbate Peroxidase and Nudix Hydrolase in Arabidopsis. In Redox State as a Central Regulator of Plant-Cell Stress Responses; Springer: Cham, Switzerland, 2016; pp. 51–70. [Google Scholar]
- Nakano, Y.; Asada, K. Hydrogen Peroxide is Scavenged by Ascorbate-specific Peroxidase in Spinach Chloroplasts. Plant Cell Physiol. 1981, 22, 867–880. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Bhuyan, M.H.M.B.; Anee, T.I.; Parvin, K.; Nahar, K.; Mahmud, J.A.; Fujita, M. Regulation of Ascorbate-Glutathione Pathway in Mitigating Oxidative Damage in Plants under Abiotic Stress. Antioxidants 2019, 8, 384. [Google Scholar] [CrossRef] [Green Version]
- Krasnovsky, A.A. Singlet molecular oxygen in photobiochemical systems: IR phosphorescence studies. Membr. Cell Biol. 1998, 12, 665–690. [Google Scholar]
- Foyer, C.H.; Noctor, G. Redox homeostasis and antioxidant signaling: A metabolic interface between stress perception and physiological responses. Plant Cell 2005, 17, 1866–1875. [Google Scholar] [CrossRef] [Green Version]
- Mosblech, A.; Feussner, I.; Heilmann, I. Oxylipins: Structurally diverse metabolites from fatty acid oxidation. Plant Physiol. Biochem. 2009, 47, 511–517. [Google Scholar] [CrossRef]
- Böttcher, C.; Pollmann, S. Plant oxylipins: Plant responses to 12-oxo-phytodienoic acid are governed by its specific structural and functional properties. FEBS J. 2009, 276, 4693–4704. [Google Scholar] [CrossRef] [Green Version]
- Yan, Y.; Borrego, E.; Kolomiets, M.V. Jasmonate Biosynthesis, Perception and Function in Plant Development and Stress Responses. In Lipid Metabolism; InTech: Rijeka, Croatia, 2013; pp. 393–442. [Google Scholar]
- Andersson, M.X.; Hamberg, M.; Kourtchenko, O.; Brunnström, A.; McPhail, K.L.; Gerwick, W.H.; Göbel, C.; Feussner, I.; Ellerström, M. Oxylipin profiling of the hypersensitive response in Arabidopsis thaliana. Formation of a novel oxo-phytodienoic acid-containing galactolipid, arabidopside E. J. Biol. Chem. 2006, 281, 31528–31537. [Google Scholar] [CrossRef] [Green Version]
- Smith, T.; McCracken, J.; Shin, Y.K.; DeWitt, D. Arachidonic acid and nonsteroidal anti-inflammatory drugs induce conformational changes in the human prostaglandin endoperoxide H2 synthase-2 (cyclooxygenase-2). J. Biol. Chem. 2000, 275, 40407–40415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feussner, I.; Wasternack, C. The lipoxygenase pathway. Annu. Rev. Plant Biol. 2002, 53, 275–297. [Google Scholar] [CrossRef] [PubMed]
- Hamberg, M.; Sanz, A.; Castresana, C. alpha-oxidation of fatty acids in higher plants. Identification of a pathogen-inducible oxygenase (piox) as an alpha-dioxygenase and biosynthesis of 2-hydroperoxylinolenic acid. J. Biol. Chem. 1999, 274, 24503–24513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liavonchanka, A.; Feussner, I. Lipoxygenases: Occurrence, functions and catalysis. J. Plant Physiol. 2006, 163, 348–357. [Google Scholar] [CrossRef] [PubMed]
- Ruan, J.; Zhou, Y.; Zhou, M.; Yan, J.; Khurshid, M.; Weng, W.; Cheng, J.; Zhang, K. Jasmonic Acid Signaling Pathway in Plants. Int. J. Mol. Sci. 2019, 20, 2479. [Google Scholar] [CrossRef] [Green Version]
- Ghorbel, M.; Brini, F.; Sharma, A.; Landi, M. Role of jasmonic acid in plants: The molecular point of view. Plant Cell Rep. 2021, 40, 1471–1494. [Google Scholar] [CrossRef]
- Wang, J.; Song, L.; Gong, X.; Xu, J.; Li, M. Functions of Jasmonic Acid in Plant Regulation and Response to Abiotic Stress. Int. J. Mol. Sci. 2020, 21, 1446. [Google Scholar] [CrossRef] [Green Version]
- Göbel, C.; Feussner, I. Methods for the analysis of oxylipins in plants. Phytochemistry 2009, 70, 1485–1503. [Google Scholar] [CrossRef]
- Collado-González, J.; Durand, T.; Ferreres, F.; Medina, S.; Torrecillas, A.; Gil-Izquierdo, Á. Phytoprostanes. Lipid Technol. 2015, 27, 127–130. [Google Scholar] [CrossRef]
- Mueller, M.J. Archetype signals in plants: The phytoprostanes. Curr. Opin. Plant Biol. 2004, 7, 441–448. [Google Scholar] [CrossRef]
- Esterbauer, H.; Schaur, R.J.; Zollner, H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free. Radic. Biol. Med. 1991, 11, 81–128. [Google Scholar] [CrossRef]
- Thoma, I.; Loeffler, C.; Sinha, A.K.; Gupta, M.; Krischke, M.; Steffan, B.; Roitsch, T.; Mueller, M.J. Cyclopentenone isoprostanes induced by reactive oxygen species trigger defense gene activation and phytoalexin accumulation in plants. Plant J. 2003, 34, 363–375. [Google Scholar] [CrossRef]
- Findling, S.; Stotz, H.U.; Zoeller, M.; Krischke, M.; Zander, M.; Gatz, C.; Berger, S.; Mueller, M.J. TGA2 signaling in response to reactive electrophile species is not dependent on cysteine modification of TGA2. PLoS ONE 2018, 13, e0195398. [Google Scholar] [CrossRef] [Green Version]
- Knieper, M.; Vogelsang, L.; Guntelmann, T.; Sproß, J.; Gröger, H.; Viehhauser, A.; Dietz, K.-J. OPDAylation of Thiols of the Redox Regulatory Network In Vitro. Antioxidants 2022, 11, 855. [Google Scholar] [CrossRef]
- Davoine, C.; Falletti, O.; Douki, T.; Iacazio, G.; Ennar, N.; Montillet, J.-L.; Triantaphylidès, C. Adducts of Oxylipin Electrophiles to Glutathione Reflect a 13 Specificity of the Downstream Lipoxygenase Pathway in the Tobacco Hypersensitive Response. Plant Physiol. 2006, 140, 1484–1493. [Google Scholar] [CrossRef] [Green Version]
- Farmer, E.E.; Davoine, C. Reactive electrophile species. Curr. Opin. Plant Biol. 2007, 10, 380–386. [Google Scholar] [CrossRef]
- Stotz, H.U.; Mueller, S.; Zoeller, M.; Mueller, M.J.; Berger, S. TGA transcription factors and jasmonate-independent COI1 signalling regulate specific plant responses to reactive oxylipins. J. Exp. Bot. 2013, 64, 963–975. [Google Scholar] [CrossRef]
- Mueller, M.J.; Berger, S. Reactive electrophilic oxylipins: Pattern recognition and signalling. Phytochemistry 2009, 70, 1511–1521. [Google Scholar] [CrossRef]
- Del Rio, D.; Stewart, A.J.; Pellegrini, N. A review of recent studies on malondialdehyde as toxic molecule and biological marker of oxidative stress. Nutr. Metab. Cardiovasc. Dis. NMCD 2005, 15, 316–328. [Google Scholar] [CrossRef]
- Farmer, E.E.; Mueller, M.J. ROS-mediated lipid peroxidation and RES-activated signaling. Annu. Rev. Plant Biol. 2013, 64, 429–450. [Google Scholar] [CrossRef]
- Ahmed, O.S.; Galano, J.-M.; Pavlickova, T.; Revol-Cavalier, J.; Vigor, C.; Lee, J.C.-Y.; Oger, C.; Durand, T. Moving forward with isoprostanes, neuroprostanes and phytoprostanes: Where are we now? Essays Biochem. 2020, 64, 463–484. [Google Scholar] [CrossRef] [PubMed]
- Thoma, I.; Krischke, M.; Loeffler, C.; Mueller, M.J. The isoprostanoid pathway in plants. Chem. Phys. Lipids 2004, 128, 135–148. [Google Scholar] [CrossRef] [PubMed]
- Baillie, T.A.; Slatter, J.G. Glutathione: A vehicle for the transport of chemically reactive metabolites in vivo. Acc. Chem. Res. 1991, 24, 264–270. [Google Scholar] [CrossRef]
- Taki, N.; Sasaki-Sekimoto, Y.; Obayashi, T.; Kikuta, A.; Kobayashi, K.; Ainai, T.; Yagi, K.; Sakurai, N.; Suzuki, H.; Masuda, T.; et al. 12-oxo-phytodienoic acid triggers expression of a distinct set of genes and plays a role in wound-induced gene expression in Arabidopsis. Plant Physiol. 2005, 139, 1268–1283. [Google Scholar] [CrossRef] [Green Version]
- Ohkama-Ohtsu, N.; Sasaki-Sekimoto, Y.; Oikawa, A.; Jikumaru, Y.; Shinoda, S.; Inoue, E.; Kamide, Y.; Yokoyama, T.; Hirai, M.Y.; Shirasu, K.; et al. 12-oxo-phytodienoic acid-glutathione conjugate is transported into the vacuole in Arabidopsis. Plant Cell Physiol. 2011, 52, 205–209. [Google Scholar] [CrossRef]
- Scala, A.; Allmann, S.; Mirabella, R.; Haring, M.A.; Schuurink, R.C. Green leaf volatiles: A plant’s multifunctional weapon against herbivores and pathogens. Int. J. Mol. Sci. 2013, 14, 17781–17811. [Google Scholar] [CrossRef] [Green Version]
- Mirabella, R.; Rauwerda, H.; Struys, E.A.; Jakobs, C.; Triantaphylidès, C.; Haring, M.A.; Schuurink, R.C. The Arabidopsis her1 mutant implicates GABA in E-2-hexenal responsiveness. Plant J. 2008, 53, 197–213. [Google Scholar] [CrossRef]
- Fujita, M.; Hossain, M.Z. Modulation of pumpkin glutathione S-transferases by aldehydes and related compounds. Plant Cell Physiol. 2003, 44, 481–490. [Google Scholar] [CrossRef] [Green Version]
- Vollenweider, S.; Weber, H.; Stolz, S.; Chételat, A.; Farmer, E.E. Fatty acid ketodienes and fatty acid ketotrienes: Michael addition acceptors that accumulate in wounded and diseased Arabidopsis leaves. Plant J. 2000, 24, 467–476. [Google Scholar] [CrossRef]
- Wasternack, C.; Feussner, I. The Oxylipin Pathways: Biochemistry and Function. Annu. Rev. Plant Biol. 2018, 69, 363–386. [Google Scholar] [CrossRef]
- Yoeun, S.; Cho, K.; Han, O. Structural Evidence for the Substrate Channeling of Rice Allene Oxide Cyclase in Biologically Analogous Nazarov Reaction. Front. Chem. 2018, 6, 500. [Google Scholar] [CrossRef]
- Vick, B.A.; Zimmerman, D.C. Characterization of 12-oxo-phytodienoic Acid reductase in corn: The jasmonic Acid pathway. Plant Physiol. 1986, 80, 202–205. [Google Scholar] [CrossRef] [Green Version]
- Staswick, P.E.; Tiryaki, I. The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell 2004, 16, 2117–2127. [Google Scholar] [CrossRef] [Green Version]
- Acosta, I.F.; Farmer, E.E. Jasmonates. Arab. Book 2010, 8, e0129. [Google Scholar] [CrossRef]
- Tang, G.; Ma, J.; Hause, B.; Nick, P.; Riemann, M. Jasmonate is required for the response to osmotic stress in rice. Environ. Exp. Bot. 2020, 175, 104047. [Google Scholar] [CrossRef]
- Chini, A.; Monte, I.; Zamarreño, A.M.; Hamberg, M.; Lassueur, S.; Reymond, P.; Weiss, S.; Stintzi, A.; Schaller, A.; Porzel, A.; et al. An OPR3-independent pathway uses 4,5-didehydrojasmonate for jasmonate synthesis. Nat. Chem. Biol. 2018, 14, 171–178. [Google Scholar] [CrossRef] [Green Version]
- Monte, I.; Kneeshaw, S.; Franco-Zorrilla, J.M.; Chini, A.; Zamarreño, A.M.; García-Mina, J.M.; Solano, R. An Ancient COI1-Independent Function for Reactive Electrophilic Oxylipins in Thermotolerance. Curr. Biol. 2020, 30, 962–971.e3. [Google Scholar] [CrossRef]
- Buseman, C.M.; Tamura, P.; Sparks, A.A.; Baughman, E.J.; Maatta, S.; Zhao, J.; Roth, M.R.; Esch, S.W.; Shah, J.; Williams, T.D.; et al. Wounding stimulates the accumulation of glycerolipids containing oxophytodienoic acid and dinor-oxophytodienoic acid in Arabidopsis leaves. Plant Physiol. 2006, 142, 28–39. [Google Scholar] [CrossRef] [Green Version]
- Dave, A.; Graham, I.A. Oxylipin Signaling: A Distinct Role for the Jasmonic Acid Precursor cis-(+)-12-Oxo-Phytodienoic Acid (cis-OPDA). Front. Plant Sci. 2012, 3, 42. [Google Scholar] [CrossRef] [Green Version]
- Böttcher, C.; Weiler, E.W. cyclo-Oxylipin-galactolipids in plants: Occurrence and dynamics. Planta 2007, 226, 629–637. [Google Scholar] [CrossRef]
- Hartley, S.E.; Eschen, R.; Horwood, J.M.; Gange, A.C.; Hill, E.M. Infection by a foliar endophyte elicits novel arabidopside-based plant defence reactions in its host, Cirsium arvense. New Phytol. 2015, 205, 816–827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nilsson, A.K.; Johansson, O.N.; Fahlberg, P.; Kommuri, M.; Töpel, M.; Bodin, L.J.; Sikora, P.; Modarres, M.; Ekengren, S.; Nguyen, C.T.; et al. Acylated monogalactosyl diacylglycerol: Prevalence in the plant kingdom and identification of an enzyme catalyzing galactolipid head group acylation in Arabidopsis thaliana. Plant J. 2015, 84, 1152–1166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nilsson, A.K.; Fahlberg, P.; Ellerström, M.; Andersson, M.X. Oxo-phytodienoic acid (OPDA) is formed on fatty acids esterified to galactolipids after tissue disruption in Arabidopsis thaliana. FEBS Lett. 2012, 586, 2483–2487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kourtchenko, O.; Andersson, M.X.; Hamberg, M.; Brunnström, A.; Göbel, C.; McPhail, K.L.; Gerwick, W.H.; Feussner, I.; Ellerström, M. Oxo-phytodienoic acid-containing galactolipids in Arabidopsis: Jasmonate signaling dependence. Plant Physiol. 2007, 145, 1658–1669. [Google Scholar] [CrossRef] [Green Version]
- Stelmach, B.A.; Müller, A.; Hennig, P.; Gebhardt, S.; Schubert-Zsilavecz, M.; Weiler, E.W. A novel class of oxylipins, sn1-O-(12-oxophytodienoyl)-sn2-O-(hexadecatrienoyl)-monogalactosyl Diglyceride, from Arabidopsis thaliana. J. Biol. Chem. 2001, 276, 12832–12838. [Google Scholar] [CrossRef] [Green Version]
- Ellinger, D.; Stingl, N.; Kubigsteltig, I.I.; Bals, T.; Juenger, M.; Pollmann, S.; Berger, S.; Schuenemann, D.; Mueller, M.J. DONGLE and DEFECTIVE IN ANTHER DEHISCENCE1 lipases are not essential for wound- and pathogen-induced jasmonate biosynthesis: Redundant lipases contribute to jasmonate formation. Plant Physiol. 2010, 153, 114–127. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Guo, Q.; Froehlich, J.E.; Hersh, H.L.; Zienkiewicz, A.; Howe, G.A.; Benning, C. Two Abscisic Acid-Responsive Plastid Lipase Genes Involved in Jasmonic Acid Biosynthesis in Arabidopsis thaliana. Plant Cell 2018, 30, 1006–1022. [Google Scholar] [CrossRef] [Green Version]
- Hisamatsu, Y.; Goto, N.; Hasegawa, K.; Shigemori, H. Senescence-promoting effect of arabidopside A. Z. Naturforsch. C J. Biosci. 2006, 61, 363–366. [Google Scholar] [CrossRef] [Green Version]
- Gronnier, J.; Germain, V.; Gouguet, P.; Cacas, J.-L.; Mongrand, S. GIPC: Glycosyl Inositol Phospho Ceramides, the major sphingolipids on earth. Plant Signal. Behav. 2016, 11, e1152438. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Yin, J.; Wu, J.-X.; Wang, L.-Y.; Liu, Y.; Huang, L.-Q.; Wang, R.-H.; Yao, N. Ceramides regulate defense response by binding to RbohD in Arabidopsis. Plant J. 2022, 109, 1427–1440. [Google Scholar] [CrossRef]
- Dar, T.A.; Uddin, M.; Khan, M.M.A.; Hakeem, K.R.; Jaleel, H. Jasmonates counter plant stress: A Review. Environ. Exp. Bot. 2015, 115, 49–57. [Google Scholar] [CrossRef]
- Wasternack, C. Action of jasmonates in plant stress responses and development—Applied aspects. Biotechnol. Adv. 2014, 32, 31–39. [Google Scholar] [CrossRef]
- Wasternack, C.; Hause, B. Jasmonates and octadecanoids: Signals in plant stress responses and development. Prog. Nucleic Acid Res. Mol. Biol. 2002, 72, 165–221. [Google Scholar] [CrossRef]
- Liu, W.; Park, S.-W. 12-oxo-Phytodienoic Acid: A Fuse and/or Switch of Plant Growth and Defense Responses? Front. Plant Sci. 2021, 12, 724079. [Google Scholar] [CrossRef]
- Jimenez Aleman, G.H.; Thirumalaikumar, V.P.; Jander, G.; Fernie, A.R.; Skirycz, A. OPDA, more than just a jasmonate precursor. Phytochemistry 2022, 204, 113432. [Google Scholar] [CrossRef]
- Liang, X.; Qian, R.; Wang, D.; Liu, L.; Sun, C.; Lin, X. Lipid-Derived Aldehydes: New Key Mediators of Plant Growth and Stress Responses. Biology 2022, 11, 1590. [Google Scholar] [CrossRef]
- Yamauchi, Y.; Furutera, A.; Seki, K.; Toyoda, Y.; Tanaka, K.; Sugimoto, Y. Malondialdehyde generated from peroxidized linolenic acid causes protein modification in heat-stressed plants. Plant Physiol. Biochem. 2008, 46, 786–793. [Google Scholar] [CrossRef]
- Yalcinkaya, T.; Uzilday, B.; Ozgur, R.; Turkan, I.; Mano, J. Lipid peroxidation-derived reactive carbonyl species (RCS): Their interaction with ROS and cellular redox during environmental stresses. Environ. Exp. Bot. 2019, 165, 139–149. [Google Scholar] [CrossRef]
- Mano, J. Reactive carbonyl species: Their production from lipid peroxides, action in environmental stress, and the detoxification mechanism. Plant Physiol. Biochem. 2012, 59, 90–97. [Google Scholar] [CrossRef]
- Mano, J.; Miyatake, F.; Hiraoka, E.; Tamoi, M. Evaluation of the toxicity of stress-related aldehydes to photosynthesis in chloroplasts. Planta 2009, 230, 639–648. [Google Scholar] [CrossRef]
- Winger, A.M.; Taylor, N.L.; Heazlewood, J.L.; Day, D.A.; Millar, A.H. The Cytotoxic lipid peroxidation product 4-hydroxy-2-nonenal covalently modifies a selective range of proteins linked to respiratory function in plant mitochondria. J. Biol. Chem. 2007, 282, 37436–37447. [Google Scholar] [CrossRef] [Green Version]
- Isom, A.L.; Barnes, S.; Wilson, L.; Kirk, M.; Coward, L.; Darley-Usmar, V. Modification of Cytochrome c by 4-hydroxy- 2-nonenal: Evidence for histidine, lysine, and arginine-aldehyde adducts. J. Am. Soc. Mass Spectrom. 2004, 15, 1136–1147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morales, M.; Munné-Bosch, S. Malondialdehyde: Facts and Artifacts. Plant Physiol. 2019, 180, 1246–1250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peñuelas, J.; Staudt, M. BVOCs and global change. Trends Plant Sci. 2010, 15, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Widhalm, J.R.; Jaini, R.; Morgan, J.A.; Dudareva, N. Rethinking how volatiles are released from plant cells. Trends Plant Sci. 2015, 20, 545–550. [Google Scholar] [CrossRef] [Green Version]
- Liao, P.; Ray, S.; Boachon, B.; Lynch, J.H.; Deshpande, A.; McAdam, S.; Morgan, J.A.; Dudareva, N. Cuticle thickness affects dynamics of volatile emission from petunia flowers. Nat. Chem. Biol. 2021, 17, 138–145. [Google Scholar] [CrossRef]
- Weber, H.; Vick, B.A.; Farmer, E.E. Dinor-oxo-phytodienoic acid: A new hexadecanoid signal in the jasmonate family. Proc. Natl. Acad. Sci. USA 1997, 94, 10473–10478. [Google Scholar] [CrossRef] [Green Version]
- Blée, E. Impact of phyto-oxylipins in plant defense. Trends Plant Sci. 2002, 7, 315–322. [Google Scholar] [CrossRef]
- Ghanem, M.E.; Ghars, M.A.; Frettinger, P.; Pérez-Alfocea, F.; Lutts, S.; Wathelet, J.-P.; Du Jardin, P.; Fauconnier, M.-L. Organ-dependent oxylipin signature in leaves and roots of salinized tomato plants (Solanum lycopersicum). J. Plant Physiol. 2012, 169, 1090–1101. [Google Scholar] [CrossRef]
- Fliegmann, J.; Schüler, G.; Boland, W.; Ebel, J.; Mithöfer, A. The role of octadecanoids and functional mimics in soybean defense responses. Biol. Chem. 2003, 384, 437–446. [Google Scholar] [CrossRef] [Green Version]
- Iqbal, M.; Evans, P.; Lledó, A.; Verdaguer, X.; Pericàs, M.A.; Riera, A.; Loeffler, C.; Sinha, A.K.; Mueller, M.J. Total synthesis and biological activity of 13,14-dehydro-12-oxo-phytodienoic acids (deoxy-J1-phytoprostanes). Chembiochem A Eur. J. Chem. Biol. 2005, 6, 276–280. [Google Scholar] [CrossRef]
- Tamogami, S.; Kodama, O. Coronatine elicits phytoalexin production in rice leaves (Oryza sativa L.) in the same manner as jasmonic acid. Phytochemistry 2000, 54, 689–694. [Google Scholar] [CrossRef]
- Kramell, R.; Miersch, O.; Atzorn, R.; Parthier, B.; Wasternack, C. Octadecanoid-derived alteration of gene expression and the “oxylipin signature” in stressed barley leaves. Implications for different signaling pathways. Plant Physiol. 2000, 123, 177–188. [Google Scholar] [CrossRef] [Green Version]
- Seltmann, M.A.; Stingl, N.E.; Lautenschlaeger, J.K.; Krischke, M.; Mueller, M.J.; Berger, S. Differential Impact of Lipoxygenase 2 and Jasmonates on Natural and Stress-Induced Senescence in Arabidopsis. Plant Physiol. 2010, 152, 1940–1950. [Google Scholar] [CrossRef] [Green Version]
- Gosset, V.; Harmel, N.; Göbel, C.; Francis, F.; Haubruge, E.; Wathelet, J.-P.; Du Jardin, P.; Feussner, I.; Fauconnier, M.-L. Attacks by a piercing-sucking insect (Myzus persicae Sultzer) or a chewing insect (Leptinotarsa decemlineata Say) on potato plants (Solanum tuberosum L.) induce differential changes in volatile compound release and oxylipin synthesis. J. Exp. Bot. 2009, 60, 1231–1240. [Google Scholar] [CrossRef] [Green Version]
- Mano, J.; Nagata, M.; Okamura, S.; Shiraya, T.; Mitsui, T. Identification of oxidatively modified proteins in salt-stressed Arabidopsis: A carbonyl-targeted proteomics approach. Plant Cell Physiol. 2014, 55, 1233–1244. [Google Scholar] [CrossRef] [Green Version]
- Mueller, S.; Hilbert, B.; Dueckershoff, K.; Roitsch, T.; Krischke, M.; Mueller, M.J.; Berger, S. General detoxification and stress responses are mediated by oxidized lipids through TGA transcription factors in Arabidopsis. Plant Cell 2008, 20, 768–785. [Google Scholar] [CrossRef] [Green Version]
- Wegener, M.; Dietz, K.-J. The mutual interaction of glycolytic enzymes and RNA in post-transcriptional regulation. RNA 2022, 28, 1446–1468. [Google Scholar] [CrossRef]
- Godbole, A.; Dubey, A.K.; Reddy, P.S.; Udayakumar, M.; Mathew, M.K. Mitochondrial VDAC and hexokinase together modulate plant programmed cell death. Protoplasma 2013, 250, 875–884. [Google Scholar] [CrossRef]
- Henry, E.; Fung, N.; Liu, J.; Drakakaki, G.; Coaker, G. Beyond glycolysis: GAPDHs are multi-functional enzymes involved in regulation of ROS, autophagy, and plant immune responses. PLoS Genet. 2015, 11, e1005199. [Google Scholar] [CrossRef] [Green Version]
- Hildebrandt, T.; Knuesting, J.; Berndt, C.; Morgan, B.; Scheibe, R. Cytosolic thiol switches regulating basic cellular functions: GAPDH as an information hub? Biol. Chem. 2015, 396, 523–537. [Google Scholar] [CrossRef] [PubMed]
- Schneider, M.; Knuesting, J.; Birkholz, O.; Heinisch, J.J.; Scheibe, R. Cytosolic GAPDH as a redox-dependent regulator of energy metabolism. BMC Plant Biol. 2018, 18, 184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uchida, K.; Stadtman, E.R. Covalent attachment of 4-hydroxynonenal to glyceraldehyde-3-phosphate dehydrogenase. A possible involvement of intra- and intermolecular cross-linking reaction. J. Biol. Chem. 1993, 268, 6388–6393. [Google Scholar] [CrossRef] [PubMed]
- Sauerland, M.; Mertes, R.; Morozzi, C.; Eggler, A.L.; Gamon, L.F.; Davies, M.J. Kinetic assessment of Michael addition reactions of alpha, beta-unsaturated carbonyl compounds to amino acid and protein thiols. Free Radic. Biol. Med. 2021, 169, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Tsuchiya, Y.; Yamaguchi, M.; Chikuma, T.; Hojo, H. Degradation of glyceraldehyde-3-phosphate dehydrogenase triggered by 4-hydroxy-2-nonenal and 4-hydroxy-2-hexenal. Arch. Biochem. Biophys. 2005, 438, 217–222. [Google Scholar] [CrossRef]
- Luo, W.; Komatsu, S.; Abe, T.; Matsuura, H.; Takahashi, K. Comparative Proteomic Analysis of Wild-Type Physcomitrella Patens and an OPDA-Deficient Physcomitrella Patens Mutant with Disrupted PpAOS1 and PpAOS2 Genes after Wounding. Int. J. Mol. Sci. 2020, 21, 1417. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Li, T.; Zhang, P.; Zhao, L.; Yi, D.; Zhang, Z.; Cong, B. Insights into the Jasmonate Signaling in Basal Land Plant Revealed by the Multi-Omics Analysis of an Antarctic Moss Pohlia nutans Treated with OPDA. Int. J. Mol. Sci. 2022, 23, 13507. [Google Scholar] [CrossRef]
- Sun, Y.-H.; Hung, C.-Y.; Qiu, J.; Chen, J.; Kittur, F.S.; Oldham, C.E.; Henny, R.J.; Burkey, K.O.; Fan, L.; Xie, J. Accumulation of high OPDA level correlates with reduced ROS and elevated GSH benefiting white cell survival in variegated leaves. Sci. Rep. 2017, 7, 44158. [Google Scholar] [CrossRef] [Green Version]
- Moore, M.; Smith, A.B.; Wegener, M.; Wesemann, C.; Schmidtpott, S.; Farooq, M.A.; Ganguly, D.R.; Seidel, T.; Pogson, B.J.; Dietz, K.-J. Retrograde Control of Cytosolic Translation Targets Synthesis of Plastid Proteins and Nuclear Responses for High-Light Acclimation. bioRxiv 2021. [Google Scholar] [CrossRef]
- Stenger, D.; Gruissem, W.; Baginsky, S. Mass spectrometric identification of RNA binding proteins from dried EMSA gels. J. Proteome Res. 2004, 3, 662–664. [Google Scholar] [CrossRef]
- Maynard, D.; Viehhauser, A.; Knieper, M.; Dreyer, A.; Manea, G.; Telman, W.; Butter, F.; Chibani, K.; Scheibe, R.; Dietz, K.-J. The In Vitro Interaction of 12-Oxophytodienoic Acid and Related Conjugated Carbonyl Compounds with Thiol Antioxidants. Biomolecules 2021, 11, 457. [Google Scholar] [CrossRef]
- Meyer, A.J.; Dreyer, A.; Ugalde, J.M.; Feitosa-Araujo, E.; Dietz, K.-J.; Schwarzländer, M. Shifting paradigms and novel players in Cys-based redox regulation and ROS signaling in plants—And where to go next. Biol. Chem. 2021, 402, 399–423. [Google Scholar] [CrossRef]
- Dietz, K.-J. Synergism and antagonism in plant acclimation to abiotic stress combinations. Turk. J. Bot. 2021, 45, 587–600. [Google Scholar] [CrossRef]
- Sham, A.; Moustafa, K.; Al-Ameri, S.; Al-Azzawi, A.; Iratni, R.; AbuQamar, S. Identification of Arabidopsis candidate genes in response to biotic and abiotic stresses using comparative microarrays. PLoS ONE 2015, 10, e0125666. [Google Scholar] [CrossRef] [Green Version]
- Hao, Q.; Maret, W. Aldehydes release zinc from proteins. A pathway from oxidative stress/lipid peroxidation to cellular functions of zinc. FEBS J. 2006, 273, 4300–4310. [Google Scholar] [CrossRef]
- Yalcinkaya, T.; Uzilday, B.; Ozgur, R.; Turkan, I. The roles of reactive carbonyl species in induction of antioxidant defence and ROS signalling in extreme halophytic model Eutrema parvulum and glycophytic model Arabidopsis thaliana. Environ. Exp. Bot. 2019, 160, 81–91. [Google Scholar] [CrossRef]
- Dueckershoff, K.; Mueller, S.; Mueller, M.J.; Reinders, J. Impact of cyclopentenone-oxylipins on the proteome of Arabidopsis thaliana. Biochim. Biophys. Acta 2008, 1784, 1975–1985. [Google Scholar] [CrossRef]
- Nianiou-Obeidat, I.; Madesis, P.; Kissoudis, C.; Voulgari, G.; Chronopoulou, E.; Tsaftaris, A.; Labrou, N.E. Plant glutathione transferase-mediated stress tolerance: Functions and biotechnological applications. Plant Cell Rep. 2017, 36, 791–805. [Google Scholar] [CrossRef]
- Davoine, C.; Douki, T.; Iacazio, G.; Montillet, J.-L.; Triantaphylidès, C. Conjugation of keto fatty acids to glutathione in plant tissues. Characterization and quantification by HPLC-tandem mass spectrometry. Anal. Chem. 2005, 77, 7366–7372. [Google Scholar] [CrossRef]
- Yadav, U.C.S.; Ramana, K.V. Regulation of NF-κB-induced inflammatory signaling by lipid peroxidation-derived aldehydes. Oxidative Med. Cell. Longev. 2013, 2013, 690545. [Google Scholar] [CrossRef] [Green Version]
- Johansson, M.H. Reversible Michael additions: Covalent inhibitors and prodrugs. Mini Rev. Med. Chem. 2012, 12, 1330–1344. [Google Scholar] [CrossRef] [PubMed]
- Petersen, D.R.; Doorn, J.A. Reactions of 4-hydroxynonenal with proteins and cellular targets. Free. Radic. Biol. Med. 2004, 37, 937–945. [Google Scholar] [CrossRef]
- Esterbauer, H.; Zollner, H.; Scholz, N. Reaction of glutathione with conjugated carbonyls. Z. Naturforsch. C Biosci. 1975, 30, 466–473. [Google Scholar] [CrossRef] [PubMed]
- Missihoun, T.D.; Kotchoni, S.O.; Bartels, D. Aldehyde Dehydrogenases Function in the Homeostasis of Pyridine Nucleotides in Arabidopsis thaliana. Sci. Rep. 2018, 8, 2936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holopainen, J.K.; Gershenzon, J. Multiple stress factors and the emission of plant VOCs. Trends Plant Sci. 2010, 15, 176–184. [Google Scholar] [CrossRef]
- Riemann, M.; Dhakarey, R.; Hazman, M.; Miro, B.; Kohli, A.; Nick, P. Exploring Jasmonates in the Hormonal Network of Drought and Salinity Responses. Front. Plant Sci. 2015, 6, 1077. [Google Scholar] [CrossRef] [Green Version]
- Singh, S.; Rai, K.; Ansari, N.; Agrawal, S.B. Climate Change and Secondary Metabolism in Plants: Resilience to Disruption; Woodhead Publishing: Cambridge, UK, 2019. [Google Scholar]
- Lee, E.-J.; Kim, K.Y.; Zhang, J.; Yamaoka, Y.; Gao, P.; Kim, H.; Hwang, J.-U.; Suh, M.C.; Kang, B.; Lee, Y. Arabidopsis seedling establishment under waterlogging requires ABCG5-mediated formation of a dense cuticle layer. New Phytol. 2021, 229, 156–172. [Google Scholar] [CrossRef]
- Liu, L.; Wang, X.; Chang, C. Toward a smart skin: Harnessing cuticle biosynthesis for crop adaptation to drought, salinity, temperature, and ultraviolet stress. Front. Plant Sci. 2022, 13, 961829. [Google Scholar] [CrossRef]
- Sharma, M.; Laxmi, A. Jasmonates: Emerging Players in Controlling Temperature Stress Tolerance. Front. Plant Sci. 2015, 6, 1129. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Jiang, L.; Wang, F.; Yu, D. Jasmonate regulates the inducer of cbf expression-C-repeat binding factor/DRE binding factor1 cascade and freezing tolerance in Arabidopsis. Plant Cell 2013, 25, 2907–2924. [Google Scholar] [CrossRef] [Green Version]
- Bittner, A.; Hause, B.; Baier, M. Cold-priming causes dampening of oxylipin biosynthesis and signalling during the early cold- and light-triggering response of Arabidopsis thaliana. J. Exp. Bot. 2021, 72, 7163–7179. [Google Scholar] [CrossRef]
- Hill, J.; Becker, H.C.; Tigerstedt, P.M.A. Quantitative and Ecological Aspects of Plant Breeding; Chapman & Hall: London, UK, 1998; ISBN 9780412753909. [Google Scholar]
- Singla, J.; Krattinger, S.G. Biotic Stress Resistance Genes in Wheat. Encyclopedia of Food Grains; Elsevier: Amsterdam, The Netherlands, 2016; pp. 388–392. ISBN 9780123947864. [Google Scholar]
- Oerke, E.-C. Crop losses to pests. J. Agric. Sci. 2006, 144, 31–43. [Google Scholar] [CrossRef]
- Wang, M.; Zheng, Q.; Shen, Q.; Guo, S. The critical role of potassium in plant stress response. Int. J. Mol. Sci. 2013, 14, 7370–7390. [Google Scholar] [CrossRef] [Green Version]
- Malathi, V.M.; Lakshmi, M.A.; Charles, S. The Applications of Genomics and Transcriptomics Approaches for Biotic Stress Tolerance in Crops. In Principles and Practices of OMICS and Genome Editing for Crop Improvement; Prakash, C.S., Fiaz, S., Fahad, S., Eds.; Springer International Publishing: Cham, Switzerland, 2022; pp. 93–122. ISBN 978-3-030-96924-0. [Google Scholar]
- Savary, S.; Willocquet, L.; Pethybridge, S.J.; Esker, P.; McRoberts, N.; Nelson, A. The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 2019, 3, 430–439. [Google Scholar] [CrossRef]
- Wang, Y.; Mostafa, S.; Zeng, W.; Jin, B. Function and Mechanism of Jasmonic Acid in Plant Responses to Abiotic and Biotic Stresses. Int. J. Mol. Sci. 2021, 22, 8568. [Google Scholar] [CrossRef]
- Mostafa, S.; Wang, Y.; Zeng, W.; Jin, B. Plant Responses to Herbivory, Wounding, and Infection. Int. J. Mol. Sci. 2022, 23, 7031. [Google Scholar] [CrossRef]
- Gimenez-Ibanez, S.; Zamarreño, A.M.; García-Mina, J.M.; Solano, R. An Evolutionarily Ancient Immune System Governs the Interactions between Pseudomonas syringae and an Early-Diverging Land Plant Lineage. Curr. Biol. 2019, 29, 2270–2281.e4. [Google Scholar] [CrossRef]
- Stintzi, A.; Weber, H.; Reymond, P.; Browse, J.; Farmer, E.E. Plant defense in the absence of jasmonic acid: The role of cyclopentenones. Proc. Natl. Acad. Sci. USA 2001, 98, 12837–12842. [Google Scholar] [CrossRef] [Green Version]
- Bosch, M.; Wright, L.P.; Gershenzon, J.; Wasternack, C.; Hause, B.; Schaller, A.; Stintzi, A. Jasmonic acid and its precursor 12-oxophytodienoic acid control different aspects of constitutive and induced herbivore defenses in tomato. Plant Physiol. 2014, 166, 396–410. [Google Scholar] [CrossRef] [Green Version]
- Schulze, A.; Zimmer, M.; Mielke, S.; Stellmach, H.; Melnyk, C.W.; Hause, B.; Gasperini, D. Wound-Induced Shoot-to-Root Relocation of JA-Ile Precursors Coordinates Arabidopsis Growth. Mol. Plant 2019, 12, 1383–1394. [Google Scholar] [CrossRef] [Green Version]
- Frost, C.J.; Mescher, M.C.; Carlson, J.E.; de Moraes, C.M. Plant Defense Priming against Herbivores: Getting Ready for a Different Battle. Plant Physiol. 2008, 146, 818–824. [Google Scholar] [CrossRef] [Green Version]
- Ton, J.; D’Alessandro, M.; Jourdie, V.; Jakab, G.; Karlen, D.; Held, M.; Mauch-Mani, B.; Turlings, T.C.J. Priming by airborne signals boosts direct and indirect resistance in maize. Plant J. 2007, 49, 16–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heil, M.; Silva Bueno, J.C. Within-plant signaling by volatiles leads to induction and priming of an indirect plant defense in nature. Proc. Natl. Acad. Sci. USA 2007, 104, 5467–5472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pretorius, C.J.; Zeiss, D.R.; Dubery, I.A. The presence of oxygenated lipids in plant defense in response to biotic stress: A metabolomics appraisal. Plant Signal. Behav. 2021, 16, 1989215. [Google Scholar] [CrossRef] [PubMed]
- Desbois, A.P.; Smith, V.J. Antibacterial free fatty acids: Activities, mechanisms of action and biotechnological potential. Appl. Microbiol. Biotechnol. 2010, 85, 1629–1642. [Google Scholar] [CrossRef] [Green Version]
- Stulnig, T.M.; Huber, J.; Leitinger, N.; Imre, E.M.; Angelisova, P.; Nowotny, P.; Waldhausl, W. Polyunsaturated eicosapentaenoic acid displaces proteins from membrane rafts by altering raft lipid composition. J. Biol. Chem. 2001, 276, 37335–37340. [Google Scholar] [CrossRef] [Green Version]
- Kachroo, A.; Kachroo, P. Fatty Acid-derived signals in plant defense. Annu. Rev. Phytopathol. 2009, 47, 153–176. [Google Scholar] [CrossRef]
- Maia, M.R.G.; Chaudhary, L.C.; Bestwick, C.S.; Richardson, A.J.; McKain, N.; Larson, T.R.; Graham, I.A.; Wallace, R.J. Toxicity of unsaturated fatty acids to the biohydrogenating ruminal bacterium, Butyrivibrio fibrisolvens. BMC Microbiol. 2010, 10, 52. [Google Scholar] [CrossRef] [Green Version]
- Schieferle, S.; Tappe, B.; Korte, P.; Mueller, M.J.; Berger, S. Pathogens and Elicitors Induce Local and Systemic Changes in Triacylglycerol Metabolism in Roots and in Leaves of Arabidopsis thaliana. Biology 2021, 10, 920. [Google Scholar] [CrossRef]
- Lim, G.-H.; Singhal, R.; Kachroo, A.; Kachroo, P. Fatty Acid- and Lipid-Mediated Signaling in Plant Defense. Annu. Rev. Phytopathol. 2017, 55, 505–536. [Google Scholar] [CrossRef]
- Zheng, C.J.; Yoo, J.-S.; Lee, T.-G.; Cho, H.-Y.; Kim, Y.-H.; Kim, W.-G. Fatty acid synthesis is a target for antibacterial activity of unsaturated fatty acids. FEBS Lett. 2005, 579, 5157–5162. [Google Scholar] [CrossRef] [Green Version]
- Pieterse, C.M.J.; Zamioudis, C.; Berendsen, R.L.; Weller, D.M.; van Wees, S.C.M.; Bakker, P.A.H.M. Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 2014, 52, 347–375. [Google Scholar] [CrossRef] [Green Version]
- Carella, P. Xylem-Mobile Oxylipins Are Critical Regulators of Induced Systemic Resistance in Maize. Plant Cell 2020, 32, 13–14. [Google Scholar] [CrossRef] [Green Version]
- Constantino, N.N.; Mastouri, F.; Damarwinasis, R.; Borrego, E.J.; Moran-Diez, M.E.; Kenerley, C.M.; Gao, X.; Kolomiets, M.V. Root-expressed maize lipoxygenase 3 negatively regulates induced systemic resistance to Colletotrichum graminicola in shoots. Front. Plant Sci. 2013, 4, 510. [Google Scholar] [CrossRef] [Green Version]
- Woldemariam, M.G.; Ahern, K.; Jander, G.; Tzin, V. A role for 9-lipoxygenases in maize defense against insect herbivory. Plant Signal. Behav. 2018, 13, e1422462. [Google Scholar] [CrossRef] [Green Version]
- Djonovic, S.; Vargas, W.A.; Kolomiets, M.V.; Horndeski, M.; Wiest, A.; Kenerley, C.M. A proteinaceous elicitor Sm1 from the beneficial fungus Trichoderma virens is required for induced systemic resistance in maize. Plant Physiol. 2007, 145, 875–889. [Google Scholar] [CrossRef] [Green Version]
- Park, Y.S. Diverse functions of the two segmentally duplicated 9-lipoxygenases ZmLOX4 and ZmLOX5 of maize: Diverse functions of the two segmentally duplicated 9-lipoxygenases ZmLOX4 and ZmLOX5 of maize. Doctoral Dissertation, Texas A&M University, College Station, TX, USA, 2012. [Google Scholar]
- Christensen, S.A. The Function of the Lipoxygenase ZmLOX10 in Maize Interactions with Insects and Pathogens: The Function of the Lipoxygenase ZmLOX10 in Maize Interactions with Insects and Pathogens. Doctoral Dissertation, Texas A&M University, College Station, TX, USA, 2011. [Google Scholar]
- Wang, K.-D.; Borrego, E.J.; Kenerley, C.M.; Kolomiets, M.V. Oxylipins Other Than Jasmonic Acid Are Xylem-Resident Signals Regulating Systemic Resistance Induced by Trichoderma virens in Maize. Plant Cell 2020, 32, 166–185. [Google Scholar] [CrossRef]
- Gao, X.; Starr, J.; Göbel, C.; Engelberth, J.; Feussner, I.; Tumlinson, J.; Kolomiets, M. Maize 9-lipoxygenase ZmLOX3 controls development, root-specific expression of defense genes, and resistance to root-knot nematodes. Mol. Plant-Microbe Interact. MPMI 2008, 21, 98–109. [Google Scholar] [CrossRef] [Green Version]
- Balint-Kurti, P. The plant hypersensitive response: Concepts, control and consequences. Mol. Plant Pathol. 2019, 20, 1163–1178. [Google Scholar] [CrossRef] [Green Version]
- Goodmann, R.N.; Novacky, A.J. The Hypersensitive Reaction in Plants to Pathogens: A Resistance Phenomenon; American Phytopathological Society (APS): St. Paul, MN, USA, 1994. [Google Scholar]
- Danon, A.; Delorme, V.; Mailhac, N.; Gallois, P. Plant programmed cell death: A common way to die. Plant Physiol. Biochem. 2000, 38, 647–655. [Google Scholar] [CrossRef]
- Naveed, Z.A.; Wei, X.; Chen, J.; Mubeen, H.; Ali, G.S. The PTI to ETI Continuum in Phytophthora-Plant Interactions. Front. Plant Sci. 2020, 11, 593905. [Google Scholar] [CrossRef] [PubMed]
- Nimchuk, Z.; Eulgem, T.; Holt, B.F.; Dangl, J.L. Recognition and response in the plant immune system. Annu. Rev. Genet. 2003, 37, 579–609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Chai, J. Structural Insights into the Plant Immune Receptors PRRs and NLRs. Plant Physiol. 2020, 182, 1566–1581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bigeard, J.; Colcombet, J.; Hirt, H. Signaling mechanisms in pattern-triggered immunity (PTI). Mol. Plant 2015, 8, 521–539. [Google Scholar] [CrossRef] [Green Version]
- Wu, L.; Chen, H.; Curtis, C.; Fu, Z.Q. Go in for the kill: How plants deploy effector-triggered immunity to combat pathogens. Corrected. Virulence 2014, 5, 710–721. [Google Scholar] [CrossRef]
- Ding, L.-N.; Li, Y.-T.; Wu, Y.-Z.; Li, T.; Geng, R.; Cao, J.; Zhang, W.; Tan, X.-L. Plant Disease Resistance-Related Signaling Pathways: Recent Progress and Future Prospects. Int. J. Mol. Sci. 2022, 23, 16200. [Google Scholar] [CrossRef]
- Frederickson Matika, D.E.; Loake, G.J. Redox Regulation in Plant Immune Function. Antioxid. Redox Signal. 2014, 21, 1373–1388. [Google Scholar] [CrossRef]
- Zhang, M.; Kadota, Y.; Prodromou, C.; Shirasu, K.; Pearl, L.H. Structural basis for assembly of Hsp90-Sgt1-CHORD protein complexes: Implications for chaperoning of NLR innate immunity receptors. Mol. Cell 2010, 39, 269–281. [Google Scholar] [CrossRef]
- de Pinto, M.C.; Locato, V.; de Gara, L. Redox regulation in plant programmed cell death. Plant Cell Environ. 2012, 35, 234–244. [Google Scholar] [CrossRef] [Green Version]
- Locato, V.; de Pinto, M.C.; de Gara, L. Different involvement of the mitochondrial, plastidial and cytosolic ascorbate-glutathione redox enzymes in heat shock responses. Physiol. Plant. 2009, 135, 296–306. [Google Scholar] [CrossRef]
- Mata-Pérez, C.; Spoel, S.H. Thioredoxin-mediated redox signalling in plant immunity. Plant Sci. 2019, 279, 27–33. [Google Scholar] [CrossRef]
- Biswas, M.S.; Mano, J. Reactive Carbonyl Species Activate Caspase-3-Like Protease to Initiate Programmed Cell Death in Plants. Plant Cell Physiol. 2016, 57, 1432–1442. [Google Scholar] [CrossRef] [Green Version]
- Mehta, S.; Chakraborty, A.; Roy, A.; Singh, I.K.; Singh, A. Fight Hard or Die Trying: Current Status of Lipid Signaling during Plant-Pathogen Interaction. Plants 2021, 10, 1098. [Google Scholar] [CrossRef]
- Walley, J.W.; Kliebenstein, D.J.; Bostock, R.M.; Dehesh, K. Fatty acids and early detection of pathogens. Curr. Opin. Plant Biol. 2013, 16, 520–526. [Google Scholar] [CrossRef] [Green Version]
- Savchenko, T.; Rolletschek, H.; Heinzel, N.; Tikhonov, K.; Dehesh, K. Waterlogging tolerance rendered by oxylipin-mediated metabolic reprogramming in Arabidopsis. J. Exp. Bot. 2019, 70, 2919–2932. [Google Scholar] [CrossRef]
- Yuan, L.-B.; Dai, Y.-S.; Xie, L.-J.; Yu, L.-J.; Zhou, Y.; Lai, Y.-X.; Yang, Y.-C.; Xu, L.; Chen, Q.-F.; Xiao, S. Jasmonate Regulates Plant Responses to Postsubmergence Reoxygenation through Transcriptional Activation of Antioxidant Synthesis. Plant Physiol. 2017, 173, 1864–1880. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Komatsu, S. The Role of Phytohormones in Plant Response to Flooding. Int. J. Mol. Sci. 2022, 23, 6383. [Google Scholar] [CrossRef]
- Kumar, V.; Vogelsang, L.; Seidel, T.; Schmidt, R.; Weber, M.; Reichelt, M.; Meyer, A.; Clemens, S.; Sharma, S.S.; Dietz, K.-J. Interference between arsenic-induced toxicity and hypoxia. Plant Cell Environ. 2019, 42, 574–590. [Google Scholar] [CrossRef]
- Savchenko, T.; Kolla, V.A.; Wang, C.-Q.; Nasafi, Z.; Hicks, D.R.; Phadungchob, B.; Chehab, W.E.; Brandizzi, F.; Froehlich, J.; Dehesh, K. Functional convergence of oxylipin and abscisic acid pathways controls stomatal closure in response to drought. Plant Physiol. 2014, 164, 1151–1160. [Google Scholar] [CrossRef] [Green Version]
- Savchenko, T.; Yanykin, D.; Khorobrykh, A.; Terentyev, V.; Klimov, V.; Dehesh, K. The hydroperoxide lyase branch of the oxylipin pathway protects against photoinhibition of photosynthesis. Planta 2017, 245, 1179–1192. [Google Scholar] [CrossRef]







| ROS and Redox | Protein | TAIR | Regulation by Oxylipins | Reference | |
|---|---|---|---|---|---|
| Network | Synthesis | Activity | |||
| Redox transmitter | TRX | At5g42980 At1g45145 At3g02730 At3g15360 | ↓OPDA | [56,133] | |
| GRX | At1g28480 At5g40370 At4g28730 | ↑OPDA | ↓OPDA | [56,66] | |
| Redox sensor | PRXIIB | At1g65980 | ↓OPDA | [56] | |
| 2-CysPRX | At3g11630 | ↓OPDA | [133] | ||
| APX | At1g07890 | ↑4-HNE, acrolein | [118,138] | ||
| GPX | At4g11600 | ↑OPDA | [66] | ||
| GAPDH | At1g12900 | ↓OPDA ↑OPDA | ↓OPDA ? 4-HNE | [56,103,129] | |
| At1g79530 | |||||
| At3g04120 At1g13440 | |||||
| Redox target protein | Cyp20-3 | At3g62030 | ↑OPDA ? 4-HNE | [28,118,133] | |
| Cysteine synthase | At3g59760 | ↑OPDA | ? 4-HNE | [66,118] | |
| GSTs | At2g29450 At1g02930 At2g30860 | ↑OPDA, acrolein, MDA | ? 4-HNE | [66,71,118,119,129] | |
| HSP | At3g12580 At3g46230 At5g12020 At4g10250 At5g12030 At1g525690 | ↑OPDA, PPA1 | ? 4-HNE | [66,103,118,119,136] | |
| FBPase | At1g43670 | ↓acrolein | [102] | ||
| DHAR2 | At1g19570 | ↑OPDA | [66,129] | ||
| MDHAR | At1g63940 | ? 4-HNE | [103] | ||
| ROS synthesis | Mn SOD | At3g10920 | ? 4-HNE | [103,118] | |
| Cu/Zn SOD | At2g28190 At1g08830 At2g28190 At5g23310 | ↓OPDA | ↑ 4-HNE, acrolein ↓4-HNE, acrolein | [66,136,138] | |
| RBOHs | At5g47910 At1g64060 | ↑4-HNE, acrolein ↓4-HNE, acrolein | ↑ OPDA, 4-HNE, acrolein | [96,138] | |
| ROS scavenging | Catalase | At4g35090 At1g20630 | ↑OPDA | ↑4-HNE, acrolein | [118,129,138] |
| Peroxidase 34 | At3g49120 | ? 4-HNE | [118] | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Knieper, M.; Viehhauser, A.; Dietz, K.-J. Oxylipins and Reactive Carbonyls as Regulators of the Plant Redox and Reactive Oxygen Species Network under Stress. Antioxidants 2023, 12, 814. https://doi.org/10.3390/antiox12040814
Knieper M, Viehhauser A, Dietz K-J. Oxylipins and Reactive Carbonyls as Regulators of the Plant Redox and Reactive Oxygen Species Network under Stress. Antioxidants. 2023; 12(4):814. https://doi.org/10.3390/antiox12040814
Chicago/Turabian StyleKnieper, Madita, Andrea Viehhauser, and Karl-Josef Dietz. 2023. "Oxylipins and Reactive Carbonyls as Regulators of the Plant Redox and Reactive Oxygen Species Network under Stress" Antioxidants 12, no. 4: 814. https://doi.org/10.3390/antiox12040814