The Versatile Role of miR-21 in Renal Homeostasis and Diseases
(This article belongs to the Section Cell Microenvironment)
Abstract
:1. Introduction
2. miR-21’s Genomic Organization, Biogenesis and Regulation
3. miR-21 in Kidney Development and Normal Tissue
3.1. miR-21 in Kidney Development
3.2. miR-21 in Renal Regeneration
3.3. miR-21 in Normal Tissue
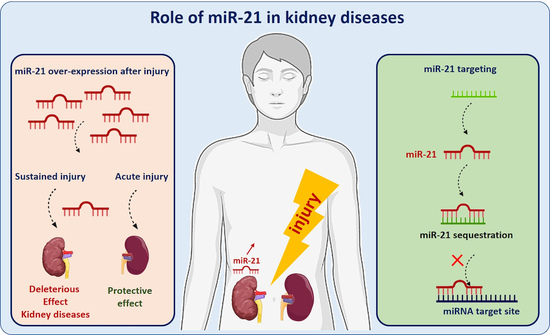
4. miR-21 in Kidney Injuries and Diseases
4.1. miR-21 in Acute and Chronic Kidney Diseases
4.1.1. A Protective Role of miR-21 in Acute Kidney Injury?
4.1.2. Sustained and Persistent Expression of miR-21 Has a Deleterious Impact in Chronic Kidney Diseases
4.2. miR-21 as an “oncomiR”
5. Potential of miR-21 as a Therapeutic Target
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Ludwig, N.; Leidinger, P.; Becker, K.; Backes, C.; Fehlmann, T.; Pallasch, C.; Rheinheimer, S.; Meder, B.; Stähler, C.; Meese, E.; et al. Distribution of miRNA expression across human tissues. Nucleic Acids Res. 2016, 44, 3865–3877. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Wightman, B.; Ha, I.; Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 1993, 75, 855–862. [Google Scholar] [CrossRef]
- Lagos-Quintana, M.; Rauhut, R.; Lendeckel, W.; Tuschl, T. Identification of novel genes coding for small expressed RNAs. Science 2001, 294, 853–858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lau, N.C.; Lim, L.P.; Weinstein, E.G.; Bartel, D.P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 2001, 294, 858–862. [Google Scholar] [CrossRef] [Green Version]
- Lee, R.C.; Ambros, V. An extensive class of small RNAs in Caenorhabditis elegans. Science 2001, 294, 862–864. [Google Scholar] [CrossRef] [Green Version]
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef] [Green Version]
- Krichevsky, A.M.; Gabriely, G. miR-21: A small multi-faceted RNA. J. Cell. Mol. Med. 2009, 13, 39–53. [Google Scholar] [CrossRef]
- Chan, J.A.; Krichevsky, A.M.; Kosik, K.S. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005, 65, 6029–6033. [Google Scholar] [CrossRef] [Green Version]
- Ribas, J.; Ni, X.; Castanares, M.; Liu, M.M.; Esopi, D.; Yegnasubramanian, S.; Rodriguez, R.; Mendell, J.T.; Lupold, S.E. A novel source for miR-21 expression through the alternative polyadenylation of VMP1 gene transcripts. Nucleic Acids Res. 2012, 40, 6821–6833. [Google Scholar] [CrossRef]
- Suster, I.; Feng, Y. Multifaceted Regulation of MicroRNA Biogenesis: Essential Roles and Functional Integration in Neuronal and Glial Development. Int. J. Mol. Sci. 2021, 22, 6765. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, P.; Heimbruch, K.E.; Rao, S. Genome-Wide Maps of Transcription Regulatory Elements and Transcription Enhancers in Development and Disease. Compr. Physiol. 2018, 9, 439–455. [Google Scholar] [CrossRef] [PubMed]
- Mudduluru, G.; George-William, J.N.; Muppala, S.; Asangani, I.A.; Kumarswamy, R.; Nelson, L.D.; Allgayer, H. Curcumin regulates miR-21 expression and inhibits invasion and metastasis in colorectal cancer. Biosci. Rep. 2011, 31, 185–197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujita, S.; Ito, T.; Mizutani, T.; Minoguchi, S.; Yamamichi, N.; Sakurai, K.; Iba, H. miR-21 Gene expression triggered by AP-1 is sustained through a double-negative feedback mechanism. J. Mol. Biol. 2008, 378, 492–504. [Google Scholar] [CrossRef]
- Cai, X.; Hagedorn, C.H.; Cullen, B.R. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 2004, 10, 1957–1966. [Google Scholar] [CrossRef] [Green Version]
- Löffler, D.; Brocke-Heidrich, K.; Pfeifer, G.; Stocsits, C.; Hackermüller, J.; Kretzschmar, A.K.; Burger, R.; Gramatzki, M.; Blumert, C.; Bauer, K.; et al. Interleukin-6 dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood 2007, 110, 1330–1333. [Google Scholar] [CrossRef] [Green Version]
- Ramalingam, P.; Palanichamy, J.K.; Singh, A.; Das, P.; Bhagat, M.; Kassab, M.A.; Sinha, S.; Chattopadhyay, P. Biogenesis of intronic miRNAs located in clusters by independent transcription and alternative splicing. RNA 2014, 20, 76–87. [Google Scholar] [CrossRef] [Green Version]
- Kumarswamy, R.; Volkmann, I.; Thum, T. Regulation and function of miRNA-21 in health and disease. RNA Biol. 2011, 8, 706–713. [Google Scholar] [CrossRef] [Green Version]
- Velu, C.S.; Baktula, A.M.; Grimes, H.L. Gfi1 regulates miR-21 and miR-196b to control myelopoiesis. Blood 2009, 113, 4720–4728. [Google Scholar] [CrossRef] [Green Version]
- Baer, C.; Claus, R.; Frenzel, L.P.; Zucknick, M.; Park, Y.J.; Gu, L.; Weichenhan, D.; Fischer, M.; Pallasch, C.P.; Herpel, E.; et al. Extensive promoter DNA hypermethylation and hypomethylation is associated with aberrant microRNA expression in chronic lymphocytic leukemia. Cancer Res. 2012, 72, 3775–3785. [Google Scholar] [CrossRef]
- Iorio, M.V.; Visone, R.; Di Leva, G.; Donati, V.; Petrocca, F.; Casalini, P.; Taccioli, C.; Volinia, S.; Liu, C.G.; Alder, H.; et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007, 67, 8699–8707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davis, B.N.; Hilyard, A.C.; Nguyen, P.H.; Lagna, G.; Hata, A. Smad proteins bind a conserved RNA sequence to promote microRNA maturation by Drosha. Mol. Cell 2010, 39, 373–384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Androsavich, J.R.; Chau, B.N.; Bhat, B.; Linsley, P.S.; Walter, N.G. Disease-linked microRNA-21 exhibits drastically reduced mRNA binding and silencing activity in healthy mouse liver. RNA 2012, 18, 1510–1526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maroney, P.A.; Yu, Y.; Fisher, J.; Nilsen, T.W. Evidence that microRNAs are associated with translating messenger RNAs in human cells. Nat. Struct. Mol. Biol. 2006, 13, 1102–1107. [Google Scholar] [CrossRef] [PubMed]
- Pickard, M.R.; Williams, G.T. Molecular and Cellular Mechanisms of Action of Tumour Suppressor GAS5 LncRNA. Genes 2015, 6, 484–499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Zhu, Z.; Watabe, K.; Zhang, X.; Bai, C.; Xu, M.; Wu, F.; Mo, Y.Y. Negative regulation of lncRNA GAS5 by miR-21. Cell Death Differ. 2013, 20, 1558–1568. [Google Scholar] [CrossRef] [Green Version]
- Poliseno, L.; Salmena, L.; Zhang, J.; Carver, B.; Haveman, W.J.; Pandolfi, P.P. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 2010, 465, 1033–1038. [Google Scholar] [CrossRef] [Green Version]
- Ebert, M.S.; Sharp, P.A. MicroRNA sponges: Progress and possibilities. RNA 2010, 16, 2043–2050. [Google Scholar] [CrossRef] [Green Version]
- Ebert, M.S.; Neilson, J.R.; Sharp, P.A. MicroRNA sponges: Competitive inhibitors of small RNAs in mammalian cells. Nat. Methods 2007, 4, 721–726. [Google Scholar] [CrossRef]
- Sayed, D.; Abdellatif, M. MicroRNAs in development and disease. Physiol. Rev. 2011, 91, 827–887. [Google Scholar] [CrossRef]
- Du, C.; Liu, C.; Kang, J.; Zhao, G.; Ye, Z.; Huang, S.; Li, Z.; Wu, Z.; Pei, G. MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat. Immunol. 2009, 10, 1252–1259, Erratum in Nat. Immunol. 2010, 11, 543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pottier, N.; Cauffiez, C.; Perrais, M.; Barbry, P.; Mari, B. FibromiRs: Translating molecular discoveries into new anti-fibrotic drugs. Trends Pharmacol. Sci. 2014, 35, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Laurent, L.C. MicroRNAs in embryonic stem cells and early embryonic development. J. Cell. Mol. Med. 2008, 12, 2181–2188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagalakshmi, V.K.; Ren, Q.; Pugh, M.M.; Valerius, M.T.; McMahon, A.P.; Yu, J. Dicer regulates the development of nephrogenic and ureteric compartments in the mammalian kidney. Kidney Int. 2011, 79, 317–330. [Google Scholar] [CrossRef] [Green Version]
- Nakagawa, N.; Xin, C.; Roach, A.M.; Naiman, N.; Shankland, S.J.; Ligresti, G.; Ren, S.; Szak, S.; Gomez, I.G.; Duffield, J.S. Dicer1 activity in the stromal compartment regulates nephron differentiation and vascular patterning during mammalian kidney organogenesis. Kidney Int. 2015, 87, 1125–1140. [Google Scholar] [CrossRef] [Green Version]
- Ho, J.; Ng, K.H.; Rosen, S.; Dostal, A.; Gregory, R.I.; Kreidberg, J.A. Podocyte-specific loss of functional microRNAs leads to rapid glomerular and tubular injury. J. Am. Soc. Nephrol. 2008, 19, 2069–2075. [Google Scholar] [CrossRef] [Green Version]
- Harvey, S.J.; Jarad, G.; Cunningham, J.; Goldberg, S.; Schermer, B.; Harfe, B.D.; McManus, M.T.; Benzing, T.; Miner, J.H. Podocyte-specific deletion of dicer alters cytoskeletal dynamics and causes glomerular disease. J. Am. Soc. Nephrol. 2008, 19, 2150–2158. [Google Scholar] [CrossRef] [Green Version]
- Sequeira-Lopez, M.L.; Weatherford, E.T.; Borges, G.R.; Monteagudo, M.C.; Pentz, E.S.; Harfe, B.D.; Carretero, O.; Sigmund, C.D.; Gomez, R.A. The microRNA-processing enzyme dicer maintains juxtaglomerular cells. J. Am. Soc. Nephrol. 2010, 21, 460–467. [Google Scholar] [CrossRef] [Green Version]
- Chen, P.Y.; Manninga, H.; Slanchev, K.; Chien, M.; Russo, J.J.; Ju, J.; Sheridan, R.; John, B.; Marks, D.S.; Gaidatzis, D.; et al. The developmental miRNA profiles of zebrafish as determined by small RNA cloning. Genes Dev. 2005, 19, 1288–1293. [Google Scholar] [CrossRef] [Green Version]
- Chau, B.N.; Xin, C.; Hartner, J.; Ren, S.; Castano, A.P.; Linn, G.; Li, J.; Tran, P.T.; Kaimal, V.; Huang, X.; et al. MicroRNA-21 promotes fibrosis of the kidney by silencing metabolic pathways. Sci. Transl. Med. 2012, 4, 121ra18. [Google Scholar] [CrossRef]
- Lai, J.Y.; Luo, J.; O’Connor, C.; Jing, X.; Nair, V.; Ju, W.; Randolph, A.; Ben-Dov, I.Z.; Matar, R.N.; Briskin, D.; et al. MicroRNA-21 in glomerular injury. J. Am. Soc. Nephrol. 2015, 26, 805–816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salice, C.J.; Rokous, J.S.; Kane, A.S.; Reimschuessel, R. New nephron development in goldfish (Carassius auratus) kidneys following repeated gentamicin-induced nephrotoxicosis. Comp. Med. 2001, 51, 56–59. [Google Scholar] [PubMed]
- Diep, C.Q.; Ma, D.; Deo, R.C.; Holm, T.M.; Naylor, R.W.; Arora, N.; Wingert, R.A.; Bollig, F.; Djordjevic, G.; Lichman, B.; et al. Identification of adult nephron progenitors capable of kidney regeneration in zebrafish. Nature 2011, 470, 95–100. [Google Scholar] [CrossRef] [Green Version]
- Hoppe, B.; Pietsch, S.; Franke, M.; Engel, S.; Groth, M.; Platzer, M.; Englert, C. MiR-21 is required for efficient kidney regeneration in fish. BMC Dev. Biol. 2015, 15, 43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patrick, D.M.; Montgomery, R.L.; Qi, X.; Obad, S.; Kauppinen, S.; Hill, J.A.; van Rooij, E.; Olson, E.N. Stress-dependent cardiac remodeling occurs in the absence of microRNA-21 in mice. J. Clin. Investig. 2010, 120, 3912–3916. [Google Scholar] [CrossRef] [Green Version]
- Hatley, M.E.; Patrick, D.M.; Garcia, M.R.; Richardson, J.A.; Bassel-Duby, R.; van Rooij, E.; Olson, E.N. Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell 2010, 18, 282–293. [Google Scholar] [CrossRef] [Green Version]
- Pickkers, P.; Ostermann, M.; Joannidis, M.; Zarbock, A.; Hoste, E.; Bellomo, R.; Prowle, J.; Darmon, M.; Bonventre, J.V.; Forni, L.; et al. The intensive care medicine agenda on acute kidney injury. Intensive Care Med. 2017, 43, 1198–1209. [Google Scholar] [CrossRef]
- Pavkovic, M.; Robinson-Cohen, C.; Chua, A.S.; Nicoara, O.; Cárdenas-González, M.; Bijol, V.; Ramachandran, K.; Hampson, L.; Pirmohamed, M.; Antoine, D.J.; et al. Detection of Drug-Induced Acute Kidney Injury in Humans Using Urinary KIM-1, miR-21, -200c, and -423. Toxicol. Sci. 2016, 152, 205–213. [Google Scholar] [CrossRef]
- Saikumar, J.; Hoffmann, D.; Kim, T.M.; Gonzalez, V.R.; Zhang, Q.; Goering, P.L.; Brown, R.P.; Bijol, V.; Park, P.J.; Waikar, S.S.; et al. Expression, circulation, and excretion profile of microRNA-21, -155, and -18a following acute kidney injury. Toxicol. Sci. 2012, 129, 256–267. [Google Scholar] [CrossRef] [Green Version]
- Du, J.; Cao, X.; Zou, L.; Chen, Y.; Guo, J.; Chen, Z.; Hu, S.; Zheng, Z. MicroRNA-21 and risk of severe acute kidney injury and poor outcomes after adult cardiac surgery. PLoS ONE 2013, 8, e63390. [Google Scholar] [CrossRef]
- Ramachandran, K.; Saikumar, J.; Bijol, V.; Koyner, J.L.; Qian, J.; Betensky, R.A.; Waikar, S.S.; Vaidya, V.S. Human miRNome profiling identifies microRNAs differentially present in the urine after kidney injury. Clin. Chem. 2013, 59, 1742–1752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Godwin, J.G.; Ge, X.; Stephan, K.; Jurisch, A.; Tullius, S.G.; Iacomini, J. Identification of a microRNA signature of renal ischemia reperfusion injury. Proc. Natl. Acad. Sci. USA 2010, 107, 14339–14344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, X.; Kriegel, A.J.; Liu, Y.; Usa, K.; Mladinov, D.; Liu, H.; Fang, Y.; Ding, X.; Liang, M. Delayed ischemic preconditioning contributes to renal protection by upregulation of miR-21. Kidney Int. 2012, 82, 1167–1175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaucsár, T.; Révész, C.; Godó, M.; Krenács, T.; Albert, M.; Szalay, C.I.; Rosivall, L.; Benyó, Z.; Bátkai, S.; Thum, T.; et al. Activation of the miR-17 family and miR-21 during murine kidney ischemia-reperfusion injury. Nucleic Acid Ther. 2013, 23, 344–354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, P.; Teng, J.; Zou, J.; Fang, Y.; Zhang, X.; Bosnjak, Z.J.; Liang, M.; Ding, X. miR-21 contributes to xenon-conferred amelioration of renal ischemia-reperfusion injury in mice. Anesthesiology 2013, 119, 621–630. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Hong, Q.; Wang, Z.; Yu, Y.; Zou, X.; Xu, L. MiR-21 inhibits autophagy by targeting Rab11a in renal ischemia/reperfusion. Exp. Cell Res. 2015, 338, 64–69. [Google Scholar] [CrossRef]
- Chen, X.; Wang, C.C.; Song, S.M.; Wei, S.Y.; Li, J.S.; Zhao, S.L.; Li, B. The administration of erythropoietin attenuates kidney injury induced by ischemia/reperfusion with increased activation of Wnt/β-catenin signaling. J. Formos. Med. Assoc. 2015, 114, 430–437. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Deng, X.; Kang, Z.; Wang, Y.; Xia, T.; Ding, N.; Yin, Y. Elevation of miR-21, through targeting MKK3, may be involved in ischemia pretreatment protection from ischemia-reperfusion induced kidney injury. J. Nephrol. 2016, 29, 27–36. [Google Scholar] [CrossRef]
- Xu, X.; Song, N.; Zhang, X.; Jiao, X.; Hu, J.; Liang, M.; Teng, J.; Ding, X. Renal Protection Mediated by Hypoxia Inducible Factor-1α Depends on Proangiogenesis Function of miR-21 by Targeting Thrombospondin 1. Transplantation 2017, 101, 1811–1819. [Google Scholar] [CrossRef] [Green Version]
- Jiao, X.; Xu, X.; Fang, Y.; Zhang, H.; Liang, M.; Teng, J.; Ding, X. miR-21 contributes to renal protection by targeting prolyl hydroxylase domain protein 2 in delayed ischaemic preconditioning. Nephrology 2017, 22, 366–373. [Google Scholar] [CrossRef]
- Pavkovic, M.; Riefke, B.; Ellinger-Ziegelbauer, H. Urinary microRNA profiling for identification of biomarkers after cisplatin-induced kidney injury. Toxicology 2014, 324, 147–157. [Google Scholar] [CrossRef] [PubMed]
- Ben-Dov, I.Z.; Muthukumar, T.; Morozov, P.; Mueller, F.B.; Tuschl, T.; Suthanthiran, M. MicroRNA sequence profiles of human kidney allografts with or without tubulointerstitial fibrosis. Transplantation 2012, 94, 1086–1094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glowacki, F.; Savary, G.; Gnemmi, V.; Buob, D.; Van der Hauwaert, C.; Lo-Guidice, J.M.; Bouyé, S.; Hazzan, M.; Pottier, N.; Perrais, M.; et al. Increased circulating miR-21 levels are associated with kidney fibrosis. PLoS ONE 2013, 8, e58014. [Google Scholar] [CrossRef] [PubMed]
- Bao, H.; Hu, S.; Zhang, C.; Shi, S.; Qin, W.; Zeng, C.; Zen, K.; Liu, Z. Inhibition of miRNA-21 prevents fibrogenic activation in podocytes and tubular cells in IgA nephropathy. Biochem. Biophys. Res. Commun. 2014, 444, 455–460. [Google Scholar] [CrossRef] [PubMed]
- McClelland, A.D.; Herman-Edelstein, M.; Komers, R.; Jha, J.C.; Winbanks, C.E.; Hagiwara, S.; Gregorevic, P.; Kantharidis, P.; Cooper, M.E. miR-21 promotes renal fibrosis in diabetic nephropathy by targeting PTEN and SMAD7. Clin. Sci. 2015, 129, 1237–1249. [Google Scholar] [CrossRef] [PubMed]
- Pezzolesi, M.G.; Satake, E.; McDonnell, K.P.; Major, M.; Smiles, A.M.; Krolewski, A.S. Circulating TGF-β1-Regulated miRNAs and the Risk of Rapid Progression to ESRD in Type 1 Diabetes. Diabetes 2015, 64, 3285–3293. [Google Scholar] [CrossRef] [Green Version]
- Zununi Vahed, S.; Omidi, Y.; Ardalan, M.; Samadi, N. Dysregulation of urinary miR-21 and miR-200b associated with interstitial fibrosis and tubular atrophy (IFTA) in renal transplant recipients. Clin. Biochem. 2017, 50, 32–39. [Google Scholar] [CrossRef]
- Hennino, M.F.; Buob, D.; Van der Hauwaert, C.; Gnemmi, V.; Jomaa, Z.; Pottier, N.; Savary, G.; Drumez, E.; Noël, C.; Cauffiez, C.; et al. miR-21-5p renal expression is associated with fibrosis and renal survival in patients with IgA nephropathy. Sci. Rep. 2016, 6, 27209. [Google Scholar] [CrossRef] [Green Version]
- Zhong, X.; Chung, A.C.; Chen, H.Y.; Meng, X.M.; Lan, H.Y. Smad3-mediated upregulation of miR-21 promotes renal fibrosis. J. Am. Soc. Nephrol. 2011, 22, 1668–1681. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Gao, Y.; Ma, M.; Li, M.; Zou, D.; Yang, J.; Zhu, Z.; Zhao, X. Effect of miR-21 on renal fibrosis by regulating MMP-9 and TIMP1 in kk-ay diabetic nephropathy mice. Cell Biochem. Biophys. 2013, 67, 537–546. [Google Scholar] [CrossRef]
- Wang, J.Y.; Gao, Y.B.; Zhang, N.; Zou, D.W.; Wang, P.; Zhu, Z.Y.; Li, J.Y.; Zhou, S.N.; Wang, S.C.; Wang, Y.Y.; et al. miR-21 overexpression enhances TGF-β1-induced epithelial-to-mesenchymal transition by target smad7 and aggravates renal damage in diabetic nephropathy. Mol. Cell. Endocrinol. 2014, 392, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Duan, L.; Tian, L.; Liu, J.; Wang, S.; Gao, Y.; Yang, J. Serum miR-21 may be a Potential Diagnostic Biomarker for Diabetic Nephropathy. Exp. Clin. Endocrinol. Diabetes 2016, 124, 417–423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, H.; Kong, L.; Tan, Y.; Epstein, P.N.; Zeng, J.; Gu, J.; Liang, G.; Kong, M.; Chen, X.; Miao, L.; et al. C66 ameliorates diabetic nephropathy in mice by both upregulating NRF2 function via increase in miR-200a and inhibiting miR-21. Diabetologia 2016, 59, 1558–1568. [Google Scholar] [CrossRef] [PubMed]
- Zarjou, A.; Yang, S.; Abraham, E.; Agarwal, A.; Liu, G. Identification of a microRNA signature in renal fibrosis: Role of miR-21. Am. J. Physiol. Ren. Physiol. 2011, 301, F793–F801. [Google Scholar] [CrossRef] [Green Version]
- Gomez, I.G.; MacKenna, D.A.; Johnson, B.G.; Kaimal, V.; Roach, A.M.; Ren, S.; Nakagawa, N.; Xin, C.; Newitt, R.; Pandya, S.; et al. Anti-microRNA-21 oligonucleotides prevent Alport nephropathy progression by stimulating metabolic pathways. J. Clin. Investig. 2015, 125, 141–156. [Google Scholar] [CrossRef]
- Huang, Y.; He, Y.; Li, J. MicroRNA-21: A central regulator of fibrotic diseases via various targets. Curr. Pharm. Des. 2015, 21, 2236–2242. [Google Scholar] [CrossRef]
- Thum, T.; Gross, C.; Fiedler, J.; Fischer, T.; Kissler, S.; Bussen, M.; Galuppo, P.; Just, S.; Rottbauer, W.; Frantz, S.; et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature 2008, 456, 980–984. [Google Scholar] [CrossRef]
- Liu, G.; Friggeri, A.; Yang, Y.; Milosevic, J.; Ding, Q.; Thannickal, V.J.; Kaminski, N.; Abraham, E. miR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J. Exp. Med. 2010, 207, 1589–1597. [Google Scholar] [CrossRef]
- Winter, J.; Jung, S.; Keller, S.; Gregory, R.I.; Diederichs, S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat. Cell Biol. 2009, 11, 228–234. [Google Scholar] [CrossRef]
- Faragalla, H.; Youssef, Y.M.; Scorilas, A.; Khalil, B.; White, N.M.; Mejia-Guerrero, S.; Khella, H.; Jewett, M.A.; Evans, A.; Lichner, Z.; et al. The clinical utility of miR-21 as a diagnostic and prognostic marker for renal cell carcinoma. J. Mol. Diagn. 2012, 14, 385–392. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014, 513, 202–209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kowalczyk, A.E.; Krazinski, B.E.; Godlewski, J.; Grzegrzolka, J.; Kiewisz, J.; Kwiatkowski, P.; Sliwinska-Jewsiewicka, A.; Dziegiel, P.; Kmiec, Z. SATB1 is Down-regulated in Clear Cell Renal Cell Carcinoma and Correlates with miR-21-5p Overexpression and Poor Prognosis. Cancer Genom. Proteom. 2016, 13, 209–217. [Google Scholar]
- Petrozza, V.; Carbone, A.; Bellissimo, T.; Porta, N.; Palleschi, G.; Pastore, A.L.; Di Carlo, A.; Della Rocca, C.; Fazi, F. Oncogenic MicroRNAs Characterization in Clear Cell Renal Cell Carcinoma. Int. J. Mol. Sci. 2015, 16, 29219–29225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaudelot, K.; Gibier, J.B.; Pottier, N.; Hémon, B.; Van Seuningen, I.; Glowacki, F.; Leroy, X.; Cauffiez, C.; Gnemmi, V.; Aubert, S.; et al. Targeting miR-21 decreases expression of multi-drug resistant genes and promotes chemosensitivity of renal carcinoma. Tumor Biol. 2017, 39, 1010428317707372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Gu, Y.; Shen, W. MicroRNA-21 functions as an oncogene and promotes cell proliferation and invasion via TIMP3 in renal cancer. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 4566–4576. [Google Scholar] [PubMed]
- Tang, K.; Xu, H. Prognostic value of meta-signature miRNAs in renal cell carcinoma: An integrated miRNA expression profiling analysis. Sci. Rep. 2015, 5, 10272. [Google Scholar] [CrossRef] [Green Version]
- Huang, M.; Zhang, T.; Yao, Z.Y.; Xing, C.; Wu, Q.; Liu, Y.W.; Xing, X.L. MicroRNA related prognosis biomarkers from high throughput sequencing data of kidney renal clear cell carcinoma. BMC Med. Genom. 2021, 14, 72. [Google Scholar] [CrossRef]
- Guo, Y.; Li, X.; Zheng, J.; Fang, J.; Pan, G.; Chen, Z. Identification of a novel immune-related microRNA prognostic model in clear cell renal cell carcinoma. Transl. Androl. Urol. 2021, 10, 888–899. [Google Scholar] [CrossRef]
- Zhao, Y.; Tao, Z.; Chen, X. Identification of the miRNA-mRNA regulatory pathways and a miR-21-5p based nomogram model in clear cell renal cell carcinoma. PeerJ 2020, 8, e10292. [Google Scholar] [CrossRef]
- Tusong, H.; Maolakuerban, N.; Guan, J.; Rexiati, M.; Wang, W.G.; Azhati, B.; Nuerrula, Y.; Wang, Y.J. Functional analysis of serum microRNAs miR-21 and miR-106a in renal cell carcinoma. Cancer Biomark. 2017, 18, 79–85. [Google Scholar] [CrossRef]
- Kalogirou, C.; Ellinger, J.; Kristiansen, G.; Hatzichristodoulou, G.; Kübler, H.; Kneitz, B.; Busch, J.; Fendler, A. Identification of miR-21-5p and miR-210-3p serum levels as biomarkers for patients with papillary renal cell carcinoma: A multicenter analysis. Transl. Androl. Urol. 2020, 9, 1314–1322. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Li, R.; Li, X.; Peng, X.; Zhang, C.; Liu, K.; Huang, G.; Lai, Y. Identification of a four-microRNA panel in serum for screening renal cell carcinoma. Pathol. Res. Pract. 2021, 227, 153625. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, J.J.; Purdue, M.P.; Signoretti, S.; Swanton, C.; Albiges, L.; Schmidinger, M.; Heng, D.Y.; Larkin, J.; Ficarra, V. Renal cell carcinoma. Nat. Rev. Dis. Prim. 2017, 3, 17009. [Google Scholar] [CrossRef] [PubMed]
- Wettersten, H.I.; Aboud, O.A.; Lara, P.N., Jr.; Weiss, R.H. Metabolic reprogramming in clear cell renal cell carcinoma. Nat. Rev. Nephrol. 2017, 13, 410–419. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Tan, T.; Zhu, L.; Dong, H.; Xian, R. Hypomethylation Causes MIR21 Overexpression in Tumors. Mol. Ther. Oncolytics 2020, 18, 47–57. [Google Scholar] [CrossRef]
- Wu, T.K.; Wei, C.W.; Pan, Y.R.; Hsu, R.J.; Wu, C.Y.; Yu, Y.L. The uremic toxin p-cresyl sulfate induces proliferation and migration of clear cell renal cell carcinoma via microRNA-21/ HIF-1α axis signals. Sci. Rep. 2019, 9, 3207. [Google Scholar] [CrossRef] [Green Version]
- An, F.; Liu, Y.; Hu, Y. miR-21 inhibition of LATS1 promotes proliferation and metastasis of renal cancer cells and tumor stem cell phenotype. Oncol. Lett. 2017, 14, 4684–4688. [Google Scholar] [CrossRef] [Green Version]
- Yuan, H.; Xin, S.; Huang, Y.; Bao, Y.; Jiang, H.; Zhou, L.; Ren, X.; Li, L.; Wang, Q.; Zhang, J. Downregulation of PDCD4 by miR-21 suppresses tumor transformation and proliferation in a nude mouse renal cancer model. Oncol. Lett. 2017, 14, 3371–3378. [Google Scholar] [CrossRef] [Green Version]
- Cao, J.; Liu, J.; Xu, R.; Zhu, X.; Liu, L.; Zhao, X. MicroRNA-21 stimulates epithelial-to-mesenchymal transition and tumorigenesis in clear cell renal cells. Mol. Med. Rep. 2016, 13, 75–82. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Xin, S.; He, Z.; Che, X.; Wang, J.; Xiao, X.; Chen, J.; Song, X. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor PDCD4 and promotes cell transformation, proliferation, and metastasis in renal cell carcinoma. Cell. Physiol. Biochem. 2014, 33, 1631–1642. [Google Scholar] [CrossRef]
- Bera, A.; Ghosh-Choudhury, N.; Dey, N.; Das, F.; Kasinath, B.S.; Abboud, H.E.; Choudhury, G.G. NFκB-mediated cyclin D1 expression by microRNA-21 influences renal cancer cell proliferation. Cell. Signal. 2013, 25, 2575–2586. [Google Scholar] [CrossRef] [PubMed]
- Bera, A.; Das, F.; Ghosh-Choudhury, N.; Kasinath, B.S.; Abboud, H.E.; Choudhury, G.G. microRNA-21-induced dissociation of PDCD4 from rictor contributes to Akt-IKKβ-mTORC1 axis to regulate renal cancer cell invasion. Exp. Cell Res. 2014, 328, 99–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dey, N.; Das, F.; Ghosh-Choudhury, N.; Mandal, C.C.; Parekh, D.J.; Block, K.; Kasinath, B.S.; Abboud, H.E.; Choudhury, G.G. microRNA-21 governs TORC1 activation in renal cancer cell proliferation and invasion. PLoS ONE 2012, 7, e37366. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Guo, Y.; Shang, C.; Song, Y.; Wu, B. miR-21 downregulated TCF21 to inhibit KISS1 in renal cancer. Urology 2012, 80, 1298–1302.e1. [Google Scholar] [CrossRef] [PubMed]
- Naro, Y.; Ankenbruck, N.; Thomas, M.; Tivon, Y.; Connelly, C.M.; Gardner, L.; Deiters, A. Small Molecule Inhibition of MicroRNA miR-21 Rescues Chemosensitivity of Renal-Cell Carcinoma to Topotecan. J. Med. Chem. 2018, 61, 5900–5909. [Google Scholar] [CrossRef]
- Liu, L.; Pang, X.; Shang, W.; Xie, H.; Feng, Y.; Feng, G. Long non-coding RNA GAS5 sensitizes renal cell carcinoma to sorafenib via miR-21/SOX5 pathway. Cell Cycle 2019, 18, 257–263. [Google Scholar] [CrossRef]
- Goujon, M.; Woszczyk, J.; Gaudelot, K.; Swierczewski, T.; Fellah, S.; Gibier, J.B.; Van Seuningen, I.; Larrue, R.; Cauffiez, C.; Gnemmi, V.; et al. A Double-Negative Feedback Interaction between miR-21 and PPAR-α in Clear Renal Cell Carcinoma. Cancers 2022, 14, 795. [Google Scholar] [CrossRef]
- Zamecnik, P.C.; Stephenson, M.L. Inhibition of Rous sarcoma virus replication and cell transformation by a specific oligodeoxynucleotide. Proc. Natl. Acad. Sci. USA 1978, 75, 280–284. [Google Scholar] [CrossRef] [Green Version]
- Fellah, S.; Larrue, R.; Truchi, M.; Vassaux, G.; Mari, B.; Cauffiez, C.; Pottier, N. Pervasive role of the long noncoding RNA DNM3OS in development and diseases. Wiley Interdiscip. Rev. RNA 2022, e1736. [Google Scholar] [CrossRef]
- Gomez, I.G.; Nakagawa, N.; Duffield, J.S. MicroRNAs as novel therapeutic targets to treat kidney injury and fibrosis. Am. J. Physiol. Ren. Physiol. 2016, 310, F931–F944. [Google Scholar] [CrossRef] [Green Version]
- Kölling, M.; Kaucsar, T.; Schauerte, C.; Hübner, A.; Dettling, A.; Park, J.K.; Busch, M.; Wulff, X.; Meier, M.; Scherf, K.; et al. Therapeutic miR-21 Silencing Ameliorates Diabetic Kidney Disease in Mice. Mol. Ther. 2017, 25, 165–180. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Larrue, R.; Fellah, S.; Van der Hauwaert, C.; Hennino, M.-F.; Perrais, M.; Lionet, A.; Glowacki, F.; Pottier, N.; Cauffiez, C. The Versatile Role of miR-21 in Renal Homeostasis and Diseases. Cells 2022, 11, 3525. https://doi.org/10.3390/cells11213525
Larrue R, Fellah S, Van der Hauwaert C, Hennino M-F, Perrais M, Lionet A, Glowacki F, Pottier N, Cauffiez C. The Versatile Role of miR-21 in Renal Homeostasis and Diseases. Cells. 2022; 11(21):3525. https://doi.org/10.3390/cells11213525
Chicago/Turabian StyleLarrue, Romain, Sandy Fellah, Cynthia Van der Hauwaert, Marie-Flore Hennino, Michaël Perrais, Arnaud Lionet, François Glowacki, Nicolas Pottier, and Christelle Cauffiez. 2022. "The Versatile Role of miR-21 in Renal Homeostasis and Diseases" Cells 11, no. 21: 3525. https://doi.org/10.3390/cells11213525