The Rhizosphere Functional Microbial Community: A Key Driver of Phosphorus Utilization Efficiency in Karst Forest Plants
Abstract
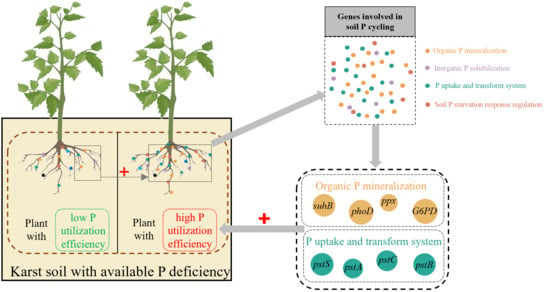
:1. Introduction
2. Material and Methods
2.1. Study Site Description
2.2. Plant Survey and Plant Nutrients Analysis
2.3. Soil Sampling and Analyses
2.4. Soil DNA Extraction and Metagenomic Sequencing
2.5. Quality Control and Genome Assembly
2.6. Bioinformatic and Functional Analysis
2.7. Statistical Analysis
3. Results
3.1. The Relationships between the Plant P Utilization Efficiency Index (PPEI) and Soil Properties
3.2. The Response of PPEI to Functional Genes Involved in Soil P Cycling
3.2.1. The Relative Abundance and Diversity of Genes Involved in Soil P Cycling
3.2.2. The Response of the PPEI to the Relative Abundance and Diversity of Genes Involved in Soil P Cycling
3.2.3. The Composition of Genes Involved in Soil P Cycling
3.2.4. The Response of the PPEI to the Four Categories of Genes Involved in Soil P Cycling
3.3. The Response of the PPEI to Genes Involved in Soil Organic P Mineralization
3.3.1. The Composition of Genes Involved in Soil Organic P Mineralization
3.3.2. The Response of the PPEI to the Six Sub-Categories of Genes Involved in Soil Organic P Mineralization
3.3.3. The Response of the PPEI to the Genes Involved in the Production of Phosphomonoesterase and Organic Pyrophosphatase
3.4. The Response of the PPEI to Genes Involved in the Soil P Uptake and Transport System
3.4.1. The Composition of Genes Involved in the Soil P Uptake and Transport System
3.4.2. The Response of the PPEI to the Four Sub-Categories of Genes Involved in the Soil P Uptake and Transport System
3.4.3. The Response of the PPEI to the Genes Involved in the Soil Phosphate Transport System
3.5. Taxonomic Composition and Contributions of Genes Involved in Soil P Cycling
3.6. The Direct Driving Factors of the PPEI
4. Discussion
4.1. Nutrient Contents of Karst Plants
4.2. The Growth of Karst Plants Was Primarily Limited by P
4.3. The Relative Abundances of Rhizosphere Functional Genes Involved in P Cycling Varied across Different Plants
4.4. Plant Rhizosphere P Cycling Microbial Community Has a Significant Positive Effect on the Plant P Utilization Efficiency (PPEI)
4.5. The Specific P Cycling Genes Involved in the Alleviation of the P Limitation for Karst Plants
4.5.1. Genes Involved in Soil Organic P Mineralization
4.5.2. Genes Involved in the Soil P Uptake and Transport System
4.6. Microbial Taxa Involved in Soil P Cycling
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vance, C.P.; Uhde-Stone, C.; Allan, D.L. Phosphorus acquisition and use: Critical adaptations by plants for securing a nonrenewable resource. New Phytol. 2003, 157, 423–447. [Google Scholar] [CrossRef]
- Huang, J.; Su, Z.; Xu, Y. The evolution of microbial phosphonate degradative pathways. J. Mol. Evol. 2005, 61, 682–690. [Google Scholar] [CrossRef]
- Peñuelas, J.; Fernández-Martínez, M.; Ciais, P.; Jou, D.; Piao, S.; Obersteiner, M.; Vicca, S.; Janssens, I.A.; Sardans, J. The bioelements, the elementome, and the biogeochemical niche. Ecology 2019, 100, e02652. [Google Scholar] [CrossRef] [PubMed]
- Menezes-Blackburn, D.; Giles, C.; Darch, T.; George, T.S.; Blackwell, M.; Stutter, M.; Shand, C.; Lumsdon, D.; Cooper, P.; Wendler, R.; et al. Opportunities for mobilizing recalcitrant phosphorus from agricultural soils: A review. Plant Soil 2017, 427, 5–16. [Google Scholar] [CrossRef] [PubMed]
- Elser, J.J.; Bracken, M.E.S.; Cleland, E.E.; Gruner, D.S.; Harpole, W.S.; Hillebrand, H.; Ngai, J.T.; Seabloom, E.W.; Shurin, J.B.; Smith, J.E. Global analysis of nitrogen and phosphorus limitation of primary producers in freshwater, marine and terrestrial ecosystems. Ecol. Lett. 2007, 10, 1135–1142. [Google Scholar] [CrossRef]
- Camenzind, T.; Hättenschwiler, S.; Treseder, K.K.; Lehmann, A.; Rillig, M.C. Nutrient limitation of soil microbial processes in tropical forests. Ecol. Monogr. 2018, 88, 4–21. [Google Scholar] [CrossRef]
- Fleischer, K.; Rammig, A.; De Kauwe, M.G.; Walker, A.P.; Domingues, T.F.; Fuchslueger, L.; Garcia, S.; Goll, D.S.; Grandis, A.; Jiang, M.; et al. Amazon forest response to CO2 fertilization dependent on plant phosphorus acquisition. Nat. Geosci. 2019, 12, 736–741. [Google Scholar] [CrossRef]
- Schneider, K.D.; Martens, J.R.T.; Zvomuya, F.; Reid, D.K.; Fraser, T.D.; Lynch, D.H.; O’Halloran, I.P.; Wilson, H.F. Options for Improved Phosphorus Cycling and Use in Agriculture at the Field and Regional Scales. J. Environ. Qual. 2019, 48, 1247–1264. [Google Scholar] [CrossRef]
- Clarholm, M.; Skyllberg, U.; Rosling, A. Organic acid induced release of nutrients from metal-stabilized soil organic matter—The unbutton model. Soil Biol. Biochem. 2015, 84, 168–176. [Google Scholar] [CrossRef]
- Pradhan, A.; Pahari, A.; Mohapatra, S.; Mishra, B.B. Phosphate-solubilizing microorganisms in sustainable agriculture: Genetic mechanism and application. In Advances in Soil Microbiology: Recent Trends and Future Prospects; Adhya, T., Mishra, B., Annapurna, K., Verma, D., Kumar, U., Eds.; Springer: Singapore, 2017; pp. 81–97. [Google Scholar]
- Liu, X.; Burslem, D.F.R.P.; Taylor, J.D.; Taylor, A.F.S.; Khoo, E.; Majalap-Lee, N.; Helgason, T.; Johnson, D. Partitioning of soil phosphorus among arbuscular and ectomycorrhizal trees in tropical and subtropical forests. Ecol. Lett. 2018, 21, 713–723. [Google Scholar] [CrossRef]
- Dai, Z.; Liu, G.; Chen, H.; Chen, C.; Wang, J.; Ai, S.; Wei, D.; Li, D.; Ma, B.; Tang, C.; et al. Long-term nutrient inputs shift soil microbial functional profiles of phosphorus cycling in diverse agroecosystems. ISME J. 2020, 14, 757–770. [Google Scholar] [CrossRef]
- Jones, D.L. Organic acids in the rhizosphere—A critical review. Plant Soil 1998, 205, 24–25. [Google Scholar] [CrossRef]
- Oburger, E.; Jones, D.L.; Wenzel, W.W. Phosphorus saturation and pH differentially regulate the efficiency of organic acid anion-mediated P solubilization mechanisms in soil. Plant Soil 2011, 341, 363–382. [Google Scholar] [CrossRef]
- Wang, Y.; Whalen, J.K.; Chen, X.; Cao, Y.; Huang, B.; Lu, C.; Shi, Y. Mechanisms for altering phosphorus sorption characteristics induced by low-molecular-weight organic acids. Can. J. Soil Sci. 2016, 96, 289–298. [Google Scholar] [CrossRef]
- Bergkemper, F.; Schöler, A.; Engel, M.; Lang, F.; Krüger, J.; Schloter, M.; Schulz, S. Phosphorus depletion in forest soils shapes bacterial communities towards phosphorus recycling systems. Environ. Microbiol. 2016, 18, 1988–2000. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.-L.; Liu, J.; Jia, P.; Yang, T.-T.; Zeng, Q.-W.; Zhang, S.-C.; Liao, B.; Shu, W.-S.; Li, J.-T. Novel phosphate-solubilizing bacteria enhance soil phosphorus cycling following ecological restoration of land degraded by mining. ISME J. 2020, 14, 1600–1613. [Google Scholar] [CrossRef] [PubMed]
- Rawat, P.; Das, S.; Shankhdhar, D.; Shankhdhar, S.C. Phosphate-Solubilizing Microorganisms: Mechanism and Their Role in Phosphate Solubilization and Uptake. J. Soil Sci. Plant Nutr. 2021, 21, 49–68. [Google Scholar] [CrossRef]
- Hobbie, J.E.; Hobbie, E.A. Microbes in nature are limited by carbon and energy: The starving-survival lifestyle in soil and consequences for estimating microbial rates. Front. Microbiol. 2013, 4, 324. [Google Scholar] [CrossRef] [PubMed]
- Sasse, J.; Martinoia, E.; Northen, T. Feed your friends: Do plant exudates shape the root microbiome? Trends Plant Sci. 2018, 23, 25–41. [Google Scholar] [CrossRef]
- Sørensen, J. The rhizosphere as a habitat for soil microorganisms. In Modern Soil Microbiology; Van Elsas, J.D., Trevors, J.T., Wellington, E.M.H., Eds.; Marcel Dekker Inc.: New York, NY, USA, 1997; pp. 21–45. [Google Scholar]
- Raaijmakers, J.M.; Paulitz, T.C.; Steinberg, C.; Alabouvette, C.; Moënne-Loccoz, Y. The rhizosphere: A playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant Soil 2009, 321, 341–361. [Google Scholar] [CrossRef]
- Hu, L.; Robert, C.A.M.; Cadot, S.; Zhang, X.; Ye, M.; Li, B.; Manzo, D.; Chervet, N.; Steinger, T.; van der Heijden, M.G.A.; et al. Root exudate metabolites drive plant-soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nat. Commun. 2018, 9, 2738. [Google Scholar] [CrossRef]
- Kuzyakov, Y.; Razavi, B.S. Rhizosphere size and shape: Temporal dynamics and spatial stationarity. Soil Biol. Biochem. 2019, 135, 343–360. [Google Scholar] [CrossRef]
- Badri, D.V.; Vivanco, J.M. Regulation and function of root exudates. Plant Cell Environ. 2009, 32, 666–681. [Google Scholar] [CrossRef] [PubMed]
- Garbeva, P.; van Elsas, J.D.; van Veen, J.A. Rhizosphere microbial community and its response to plant species and soil history. Plant Soil 2008, 302, 19–32. [Google Scholar] [CrossRef]
- Ushio, M.; Wagai, R.; Balser, T.C.; Kitayama, K. Variations in the soil microbial community composition of a tropical montane forest ecosystem: Does tree species matter. Soil Biol. Biochem. 2008, 40, 2699–2702. [Google Scholar] [CrossRef]
- Lauber, C.L.; Hamady, M.; Knight, R.; Fierer, N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 2009, 75, 5111–5120. [Google Scholar] [CrossRef]
- Oh, Y.M.; Kim, M.; Lee-Cruz, L.; Lai-Hoe, A.; Go, R.; Ainuddin, N.; Rahim, R.A.; Shukor, N.; Adams, J.M. Distinctive Bacterial Communities in the Rhizoplane of Four Tropical Tree Species. Microb. Ecol. 2012, 64, 1018–1027. [Google Scholar] [CrossRef]
- Grayston, S.J.; Wang, S.; Campbell, C.D.; Edwards, A.C. Selective influence of plant species on microbial diversity in the rhizosphere. Soil Biol. Biochem. 1998, 30, 369–378. [Google Scholar] [CrossRef]
- Bardgett, R.D.; Mawdsley, J.L.; Edwards, S.; Hobbs, P.J.; Rodwell, J.S.; Davies, W.J. Plant species and nitrogen effects on soil biological properties of temperate upland grasslands. Funct. Ecol. 1999, 13, 650–660. [Google Scholar] [CrossRef]
- Berg, G.; Smalla, K. Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol. Ecol. 2009, 68, 1–13. [Google Scholar] [CrossRef]
- Berg, G.; Roskot, N.; Steidle, A.; Eberl, L.; Zock, A.; Smalla, K. Plant-dependent genotypic and phenotypic diversity of antagonistic rhizobacteria isolated from different Verticillium host plants. Appl. Environ. Microbiol. 2002, 68, 3328–3338. [Google Scholar] [CrossRef]
- Smalla, K.; Wieland, G.; Buchner, A.; Zock, A.; Parzy, J.; Kaiser, S.; Roskot, N.; Heuer, H.; Berg, G. Bulk and rhizosphere soil bacterial communities studied by denaturing gradient gel electrophoresis: Plant-dependent enrichment and seasonal shifts revealed. Appl. Environ. Microbiol. 2001, 67, 4742–4751. [Google Scholar] [CrossRef]
- Rasche, F.; Trondl, R.; Naglreiter, C.; Reichenauer, T.G.; Sessitsch, A. Chilling and cultivar type affect the diversity of bacterial endophytes colonizing sweet pepper (Capsicum anuum L.). Can. J. Microbiol. 2006, 52, 1036–1045. [Google Scholar] [CrossRef] [PubMed]
- Bezemer, T.M.; Lawson, C.S.; Hedlund, K.; Edwards, A.R.; Brook, A.J.; Igual, J.M.; Mortimer, S.R.; VAN DER Putten, W.H. Plant species and functional group effects on abiotic and microbial soil properties and plant–soil feedback responses in two grasslands. J. Ecol. 2006, 94, 893–904. [Google Scholar] [CrossRef]
- Singh, B.K.; Munro, S.; Potts, J.M.; Millard, P. Influence of grass species and soil type on rhizosphere microbial community structure in grassland soils. Appl. Soil Ecol. 2007, 36, 147–155. [Google Scholar] [CrossRef]
- Kielak, A.; Pijl, A.S.; Van Veen, J.A.; Kowalchuk, G.A. Differences in vegetation composition and plant species identity lead to only minor changes in soil-borne microbial communities in a former arable field. FEMS Microbiol. Ecol. 2008, 63, 372–382. [Google Scholar] [CrossRef]
- Landesman, W.J.; Nelson, D.M.; Fitzpatrick, M.C. Soil properties and tree species drive ß-diversity of soil bacterial communities. Soil Biol. Biochem. 2014, 76, 201–209. [Google Scholar] [CrossRef]
- Shang, R.; Li, S.; Huang, X.; Liu, W.; Lang, X.; Su, J. Effects of soil properties and plant diversity on soil microbial community composition and diversity during secondary succession. Forests 2021, 12, 805. [Google Scholar] [CrossRef]
- Sweeting, M.M. Karst in China: Its Geomorphology and Environment; Springer Science & Business Media: Berlin, Germany, 2012. [Google Scholar]
- Ahmed, Y.A.-R.; Pichler, V.; Homolák, M.; Gömöryová, E.; Nagy, D.; Pichlerová, M.; Gregor, J. High organic carbon stock in a karstic soil of the Middle-European Forest Province persists after centuries-long agroforestry management. Eur. J. For. Res. 2012, 131, 1669–1680. [Google Scholar] [CrossRef]
- Wen, L.; Li, D.; Yang, L.; Luo, P.; Chen, H.; Xiao, K.; Song, T.; Zhang, W.; He, X.; Chen, H.; et al. Rapid recuperation of soil nitrogen following agricultural abandonment in a karst area, southwest China. Biogeochemistry 2016, 129, 341–354. [Google Scholar] [CrossRef]
- Li, D.; Wen, L.; Zhang, W.; Yang, L.; Xiao, K.; Chen, H.; Wang, K. Afforestation effects on soil organic carbon and nitrogen pools modulated by lithology. For. Ecol. Manag. 2017, 400, 85–92. [Google Scholar] [CrossRef]
- Xiao, D.; Chen, Y.; He, X.; Xu, Z.; Bai, S.H.; Zhang, W.; Cheng, M.; Hu, P.; Wang, K. Temperature and precipitation significantly influence the interactions between arbuscular mycorrhizal fungi and diazotrophs in karst ecosystems. For. Ecol. Manag. 2021, 497, 119464. [Google Scholar] [CrossRef]
- Tang, M.; Liu, J.; Hou, W.; Stubbendieck, R.M.; Xiong, H.; Jin, J.; Gong, J.; Cheng, C.; Tang, X.; Liu, Y.; et al. Structural variability in the bulk soil, rhizosphere, and root endophyte fungal communities of Themeda japonica plants under different grades of karst rocky desertification. Plant Soil 2022, 475, 105–122. [Google Scholar] [CrossRef]
- Green, S.M.; Dungait, J.A.; Tu, C.; Buss, H.L.; Sanderson, N.; Hawkes, S.J.; Xing, K.; Yue, F.; Hussey, V.L.; Peng, J.; et al. Soil functions and ecosystem services research in the Chinese karst Critical Zone. Chem. Geol. 2019, 527, 119107. [Google Scholar] [CrossRef]
- Widdig, M.; Heintz-Buschart, A.; Schleuss, P.-M.; Guhr, A.; Borer, E.T.; Seabloom, E.W.; Spohn, M. Effects of nitrogen and phosphorus addition on microbial community composition and element cycling in a grassland soil. Soil Biol. Biochem. 2020, 151, 108041. [Google Scholar] [CrossRef]
- Bloom, A.J.; Chapin, F.S., III; Mooney, H.A. Resource limitation in plants—An economic analogy. Annu. Rev. Ecol. Evol. Syst. 1985, 16, 363–392. [Google Scholar] [CrossRef]
- Liu, S.; Zhang, X.; Dungait, J.A.J.; Quine, T.A.; Razavi, B.S. Rare microbial taxa rather than phoD gene abundance determine hotspots of alkaline phosphomonoesterase activity in the karst rhizosphere soil. Biol. Fertil. Soils 2021, 57, 257–268. [Google Scholar] [CrossRef]
- He, J.-S.; Fang, J.; Wang, Z.; Guo, D.; Flynn, D.F.B.; Geng, Z. Stoichiometry and large-scale patterns of leaf carbon and nitrogen in the grassland biomes of China. Oecologia 2006, 149, 115–122. [Google Scholar] [CrossRef]
- Condit, R. Tropical Forest Census Plots: Methods and Results from Barro Colorado Island, Panama and a Comparison with Other Plots; Springer Science & Business Media: New York, NY, USA, 1998. [Google Scholar]
- Chen, S.; Waghmode, T.R.; Sun, R.; Kuramae, E.E.; Hu, C.; Liu, B. Root-associated microbiomes of wheat under the combined effect of plant development and nitrogen fertilization. Microbiome 2019, 7, 136. [Google Scholar] [CrossRef]
- Pansu, M.; Gautheyrou, J. Handbook of Soil Analysis: Mineralogical, Organic and Inorganic Methods; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2007. [Google Scholar]
- Bao, S.D. Soil and Agro-Chemistrical Analysis, 3rd ed.; Agriculture Press of China: Beijing, China, 2007. [Google Scholar]
- Qiu, X.; Wang, H.; Peng, D.; Liu, X.; Yang, F.; Li, Z.; Cheng, S. Thinning drives C:N:P stoichiometry and nutrient resorption in Larix principis-rupprechtii plantations in North China. For. Ecol. Manag. 2020, 462, 117984. [Google Scholar] [CrossRef]
- Yang, D.; Mao, H.; Jin, G. Divergent responses of foliar N:P Stoichiometry during different seasons to nitrogen deposition in an old-growth temperate forest, Northeast China. Forests 2019, 10, 257. [Google Scholar] [CrossRef]
- Bremner, J.; Mulvaney, C. Nitrogen-total. In Methods of Soil Analysis; Page, A.L., Miller, R.H., Keeney, D.R., Eds.; American Society of Agronomy and Soil Science Society of America: Madison, WI, USA, 1982; pp. 595–641. [Google Scholar]
- Pribyl, D.W. A critical review of the conventional SOC to SOM conversion factor. Geoderma 2010, 156, 75–83. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, 884–890. [Google Scholar] [CrossRef]
- Li, D.; Liu, C.-M.; Luo, R.; Sadakane, K.; Lam, T.-W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, H.; Park, J.; Takagi, T. MetaGene: Prokaryotic gene finding from environmental genome shotgun sequences. Nucleic Acids Res. 2006, 34, 5623–5630. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Yu, C.; Li, Y.; Lam, T.-W.; Yiu, S.-M.; Kristiansen, K.; Wang, J. SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics 2009, 25, 1966–1967. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using diamond. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Rong, Q.; Liu, J.; Cai, Y.; Lu, Z.; Zhao, Z.; Yue, W.; Xia, J. Leaf carbon, nitrogen and phosphorus stoichiometry of Tamarix chinensis Lour. in the Laizhou Bay coastal wetland, China. Ecol. Eng. 2015, 76, 57–65. [Google Scholar] [CrossRef]
- Cao, J.; Wang, X.; Adamowski, J.F.; Biswas, A.; Liu, C.; Chang, Z.; Feng, Q. Response of leaf stoichiometry of Oxytropis ochrocephala to elevation and slope aspect. Catena 2020, 194, 104772. [Google Scholar] [CrossRef]
- Schade, J.D.; Kyle, M.; Hobbie, S.E.; Fagan, W.F.; Elser, J.J. Stoichiometric tracking of soil nutrients by a desert insect herbivore. Ecol. Appl. 2003, 6, 96–101. [Google Scholar] [CrossRef]
- Liu, R.; Zhao, H.; Zhao, X.; Drake, S. Facilitative effects of shrubs in shifting sand on soil macro-faunal community in Horqin Sand Land of Inner Mongolia, Northern China. Eur. J. Soil Biol. 2011, 47, 316–321. [Google Scholar] [CrossRef]
- Mao, R.; Chen, H.-M.; Zhang, X.-H.; Shi, F.-X.; Song, C.-C. Effects of P addition on plant C:N:P stoichiometry in an N-limited temperate wetland of Northeast China. Sci. Total Environ. 2016, 559, 1–6. [Google Scholar] [CrossRef]
- Elser, J.J.; Fagan, W.F.; Denno, R.F.; Dobberfuhl, D.R.; Folarin, A.; Huberty, A.; Interlandi, S.; Kilham, S.S.; McCauley, E.; Schulz, K.L.; et al. Nutritional constraints in terrestrial and freshwater food webs. Nature 2000, 408, 578–580. [Google Scholar] [CrossRef]
- Reich, P.B.; Oleksyn, J. Global patterns of plant leaf N and P in relation to temperature and latitude. Proc. Natl. Acad. Sci. USA 2004, 101, 11001–11006. [Google Scholar] [CrossRef]
- Han, W.X.; Fang, J.Y.; Guo, D.L.; Zhang, Y. Leaf nitrogen and phosphorus stoichiometry across 753 terrestrial plant species in China. New Phytol. 2005, 168, 377–385. [Google Scholar] [CrossRef]
- Tang, Z.; Xu, W.; Zhou, G.; Bai, Y.; Li, J.; Tang, X.; Chen, D.; Liu, Q.; Ma, W.; Xiong, G.; et al. Patterns of plant carbon, nitrogen, and phosphorus concentration in relation to productivity in China’s terrestrial ecosystems. Proc. Natl. Acad. Sci. USA 2018, 115, 4033–4038. [Google Scholar] [CrossRef] [PubMed]
- Hedin, L.O. Global organization of terrestrial plant–nutrient interactions. Proc. Natl. Acad. Sci. USA 2004, 101, 10849–10850. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; Wu, J.; Liu, W.; Yuan, Y.; Hu, L.; Cai, Q. Linkages of plant and soil C:N:P stoichiometry and their relationships to forest growth in subtropical plantations. Plant Soil 2015, 392, 127–138. [Google Scholar] [CrossRef]
- Cleveland, C.C.; Townsend, A.R.; Schmidt, S.K. Phosphorus limitation of microbial processes in moist tropical forests: Evidence from short-term laboratory incubations and field studies. Ecosystems 2002, 5, 0680–0691. [Google Scholar] [CrossRef]
- Iyamuremye, F.; Dick, R.P.; Baham, J. Organic amendments and phosphorus sorption by soils. Adv. Agron. 1996, 56, 139–185. [Google Scholar] [CrossRef]
- Han, G.; Tang, Y.; Wu, Q.; Tan, Q. Chemical and strontium isotope characterization of rainwater in karst virgin forest, Southwest China. Atmos. Environ. 2010, 44, 174–181. [Google Scholar] [CrossRef]
- Chen, Q.; Niu, B.; Hu, Y.; Luo, T.; Zhang, G. Warming and increased precipitation indirectly affect the composition and turnover of labile-fraction soil organic matter by directly affecting vegetation and microorganisms. Sci. Total. Environ. 2020, 714, 136787. [Google Scholar] [CrossRef]
- Hofmeister, J.; Mihaljevič, M.; Hošek, J.; Sádlo, J. Eutrophication of deciduous forests in the Bohemian Karst (Czech Republic): The role of nitrogen and phosphorus. For. Ecol. Manag. 2002, 169, 213–230. [Google Scholar] [CrossRef]
- Piao, H.; Liu, C.; Zhu, S.; Zhu, J. Variations fo C4 and C3 plant N:P ratios influenced by nutrient stoichiometry in limestone and sandstone areas of Guizhou. Quat. Sci. 2005, 25, 552–560. [Google Scholar] [CrossRef]
- Zhang, W.; Zhao, J.; Pan, F.; Li, D.; Chen, H.; Wang, K. Changes in nitrogen and phosphorus limitation during secondary succession in a karst region in southwest China. Plant Soil 2015, 391, 77–91. [Google Scholar] [CrossRef]
- Sterner, R.W.; Elser, J.J. Ecological Stoichiometry: The Biology of Elements from Molecules to the Biosphere; Princeton University Press: Princeton, NJ, USA, 2003. [Google Scholar]
- Zhang, L.; Bai, Y.; Han, X. Application of N:P stoichiometry to ecology studies. J. Integr. Plant Biol. 2003, 45, 1009–1018. [Google Scholar] [CrossRef]
- Baxter, I.; Dilkes, B.P. Elemental Profiles Reflect Plant Adaptations to the Environment. Science 2012, 336, 1661–1663. [Google Scholar] [CrossRef]
- Liu, F.; Liu, Y.; Wang, G.; Song, Y.; Liu, Q.; Li, D.; Mao, P.; Zhang, H. Seasonal variations of C: N: P stoichiometry and their trade-offs in different organs of Suaeda salsa in coastal wetland of Yellow River Delta, China. PLoS ONE 2015, 10, e0138169. [Google Scholar] [CrossRef]
- Wu, T.-G.; Yu, M.-K.; Wang, G.G.; Dong, Y.; Cheng, X.-R. Leaf nitrogen and phosphorus stoichiometry across forty-two woody species in Southeast China. Biochem. Syst. Ecol. 2012, 44, 255–263. [Google Scholar] [CrossRef]
- Koerselman, W.; Meuleman, A.F.M. The Vegetation N:P ratio: A new tool to detect the nature of nutrient limitation. J. Appl. Ecol. 1996, 33, 1441. [Google Scholar] [CrossRef]
- Niinemets, U.; Kull, K. Co-limitation of plant primary productivity by nitrogen and phosphorus in a species-rich wooded meadow on calcareous soils. Acta Oecol. 2005, 28, 345–356. [Google Scholar] [CrossRef]
- Vitousek, P.M.; Porder, S.; Houlton, B.Z.; Chadwick, O.A. Terrestrial phosphorus limitation: Mechanisms, implications, and nitrogen–phosphorus interactions. Ecol. Appl. 2010, 20, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Güsewell, S. N:P ratios in terrestrial plants: Variation and functional significance. New Phytol. 2004, 164, 243–266. [Google Scholar] [CrossRef] [PubMed]
- He, J.-S.; Wang, L.; Flynn, D.F.B.; Wang, X.; Ma, W.; Fang, J. Leaf nitrogen:phosphorus stoichiometry across Chinese grassland biomes. Oecologia 2008, 155, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.; Zhuang, H.; Wu, L.; Liu, Q.; Shen, G.; Berg, B.; Man, R.; Liu, C. Variation in leaf nitrogen and phosphorus stoichiometry in Picea abies across Europe: An analysis based on local observations. For. Ecol. Manag. 2011, 261, 195–202. [Google Scholar] [CrossRef]
- Sun, L.; Gong, L.; Zhu, M.L.; Xie, L.N.; Li, H.L.; Luo, Y. Leaf stoichiometric characteristics of typical desert plants their relationships to soil environmental factors in the northern margin of the Tarin basin Chinese. J. Ecol. 2017, 36, 1208–1214. [Google Scholar] [CrossRef]
- Rifat, H.; Safdar, A.; Ummay, A.; Rabia, K.; Iftikhar, A. Soil beneficial bacteria and their role in plant growth promotion: A review. Ann. Microbiol. 2010, 60, 579–598. [Google Scholar] [CrossRef]
- Raaijmakers, J.M.; Mazzola, M. Soil immune responses. Science 2016, 352, 1392–1393. [Google Scholar] [CrossRef]
- Hartmann, A.; Rothballer, M.; Schmid, M. Lorenz Hiltner, a pioneer in rhizosphere microbial ecology and soil bacteriology research. Plant Soil 2008, 312, 7–14. [Google Scholar] [CrossRef]
- Zhou, X.; Gao, D.; Liu, J.; Qiao, P.; Zhou, X.; Lu, H.; Wu, X.; Liu, D.; Jin, X.; Wu, F. Changes in rhizosphere soil microbial communities in a continuously monocropped cucumber (Cucumis sativus L.) system. Eur. J. Soil Biol. 2014, 60, 1–8. [Google Scholar] [CrossRef]
- de Vries, F.T.; Griffiths, R.I.; Knight, C.G.; Nicolitch, O.; Williams, A. Harnessing rhizosphere microbiomes for drought-resilient crop production. Science 2020, 368, 270–274. [Google Scholar] [CrossRef] [PubMed]
- Bakker, M.G.; Otto-Hanson, L.; Lange, A.; Bradeen, J.M.; Kinkel, L.L. Plant monocultures produce more antagonistic soil Streptomyces communities than high-diversity plant communities. Soil Biol. Biochem. 2013, 65, 304–312. [Google Scholar] [CrossRef]
- Schlatter, D.C.; Bakker, M.G.; Bradeen, J.M.; Kinkel, L.L. Plant community richness and microbial interactions structure bacterial communities in soil. Ecology 2015, 96, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Xiang, X.; Shen, C.; Chu, H.; Neufeld, J.D.; Walker, V.K.; Grogan, P. Vegetation-associated impacts on arctic tundra bacterial and microeukaryotic communities. Appl. Environ. Microbiol. 2015, 81, 492–501. [Google Scholar] [CrossRef] [PubMed]
- Bulgarelli, D.; Rott, M.; Schlaeppi, K.; van Themaat, E.V.L.; Ahmadinejad, N.; Assenza, F.; Rauf, P.; Huettel, B.; Reinhardt, R.; Schmelzer, E.; et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 2012, 488, 91–95. [Google Scholar] [CrossRef]
- Lundberg, D.S.; Lebeis, S.L.; Paredes, S.H.; Yourstone, S.; Gehring, J.; Malfatti, S.; Tremblay, J.; Engelbrektson, A.; Kunin, V.; Del Rio, T.G.; et al. Defining the core Arabidopsis thaliana root microbiome. Nature 2012, 488, 86–90. [Google Scholar] [CrossRef]
- Bai, B.; Liu, W.; Qiu, X.; Zhang, J.; Zhang, J.; Bai, Y. The root microbiome: Community assembly and its contributions to plant fitness. J. Integr. Plant Biol. 2022, 64, 230–243. [Google Scholar] [CrossRef]
- Berendsen, R.L.; Pieterse, C.M.J.; Bakker, P.A.H.M. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef]
- Huang, X.-F.; Chaparro, J.M.; Reardon, K.F.; Zhang, R.; Shen, Q.; Vivanco, J.M. Rhizosphere interactions: Root exudates, microbes, and microbial communities. Botany 2014, 92, 267–275. [Google Scholar] [CrossRef]
- Hiruma, K.; Gerlach, N.; Sacristan, S.; Nakano, R.T.; Hacquard, S.; Kracher, B.; Neumann, U.; Ramirez, D.; Bucher, M.; O’Connell, R.J.; et al. Root Endophyte Colletotrichum tofieldiae Confers Plant Fitness Benefits that Are Phosphate Status Dependent. Cell 2016, 165, 464–474. [Google Scholar] [CrossRef] [PubMed]
- Kwak, M.-J.; Kong, H.G.; Choi, K.; Kwon, S.-K.; Song, J.Y.; Lee, J.; Lee, P.A.; Choi, S.Y.; Seo, M.; Lee, H.J.; et al. Rhizosphere microbiome structure alters to enable wilt resistance in tomato. Nat. Biotechnol. 2018, 36, 1100–1109. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, Y.-X.; Zhang, N.; Hu, B.; Jin, T.; Xu, H.; Qin, Y.; Yan, P.; Zhang, X.; Guo, X.; et al. NRT1.1B is associated with root microbiota composition and nitrogen use in field-grown rice. Nat. Biotechnol. 2019, 37, 676–684. [Google Scholar] [CrossRef] [PubMed]
- Ravanbakhsh, M.; A Kowalchuk, G.; Jousset, A. Root-associated microorganisms reprogram plant life history along the growth–stress resistance tradeoff. ISME J. 2019, 13, 3093–3101. [Google Scholar] [CrossRef] [PubMed]
- Salas-González, I.; Reyt, G.; Flis, P.; Custódio, V.; Gopaulchan, D.; Bakhoum, N.; Dew, T.P.; Suresh, K.; Franke, R.B.; Dangl, J.L.; et al. Coordination between microbiota and root endodermis supports plant mineral nutrient homeostasis. Science 2021, 371, 143. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Chen, D.; Zang, L.; Zhang, G.; Liu, Q.; He, Y.; Ding, F.; Wang, S.; Zhou, C.; Yang, Y.; et al. Natural restoration of degraded karst vegetation shifts soil microbial phosphorus acquisition strategies. Plant Soil 2023, 490, 201–215. [Google Scholar] [CrossRef]
- Siles, J.A.; Starke, R.; Martinovic, T.; Fernandes, M.L.P.; Orgiazzi, A.; Bastida, F. Distribution of phosphorus cycling genes across land uses and microbial taxonomic groups based on metagenome and genome mining. Soil Biol. Biochem. 2022, 174, 108826. [Google Scholar] [CrossRef]
- Hui, N.; Sun, N.; Du, H.; Umair, M.; Kang, H.; Liu, X.; Romantschuk, M.; Liu, C. Karst rocky desertification does not erode ectomycorrhizal fungal species richness but alters microbial community structure. Plant Soil 2019, 445, 383–396. [Google Scholar] [CrossRef]
- Ma, B.; Zhou, X.; Zhang, Q.; Qin, M.; Hu, L.; Yang, K.; Xie, Z.; Ma, W.; Chen, B.; Feng, H.; et al. How do soil micro-organisms respond to N, P and NP additions? Application of the ecological framework of (co-)limitation by multiple resources. J. Ecol. 2019, 107, 2329–2345. [Google Scholar] [CrossRef]
- Shen, Y.; Yu, Y.; Lucas-Borja, M.E.; Chen, F.; Chen, Q.; Tang, Y. Change of soil K, N and P following forest restoration in rock outcrop rich karst area. Catena 2020, 186, 104395. [Google Scholar] [CrossRef]
- Zhang, J.; Zheng, M.; Zhang, Y.; Wang, J.; Shen, H.; Lin, Y.; Tang, X.; Hui, D.; Lambers, H.; Sardans, J.; et al. Soil phosphorus availability affects diazotroph communities during vegetation succession in lowland subtropical forests. Appl. Soil Ecol. 2021, 166, 104009. [Google Scholar] [CrossRef]
- Turner, B.L.; Baxter, R.; A Whitton, B. Seasonal phosphatase activity in three characteristic soils of the English uplands polluted by long-term atmospheric nitrogen deposition. Environ. Pollut. 2002, 120, 313–317. [Google Scholar] [CrossRef] [PubMed]
- Spohn, M.; Kuzyakov, Y. Distribution of microbial- and root-derived phosphatase activities in the rhizosphere depending on P availability and C allocation—Coupling soil zymography with 14C imaging. Soil Biol. Biochem. 2013, 67, 106–113. [Google Scholar] [CrossRef]
- Wasaki, J.; Sakaguchi, J.; Yamamura, T.; Ito, S.; Shinano, T.; Osaki, M.; Kandeler, E. P and N deficiency change the relative abundance and function of rhizosphere microorganisms during cluster root development of white lupin (Lupinus albus L.). Soil Sci. Plant Nutr. 2018, 64, 686–696. [Google Scholar] [CrossRef]
- Wei, X.; Hu, Y.; Razavi, B.S.; Zhou, J.; Shen, J.; Nannipieri, P.; Wu, J.; Ge, T. Rare taxa of alkaline phosphomonoesterase-harboring microorganisms mediate soil phosphorus mineralization. Soil Biol. Biochem. 2019, 131, 62–70. [Google Scholar] [CrossRef]
- Ragot, S.A.; Kertesz, M.A.; Mészáros, É.; Frossard, E.; Bünemann, E.K. Soil phoD and phoX alkaline phosphatase gene diversity responds to multiple environmental factors. FEMS Microbiol. Ecol. 2017, 93, fiw212. [Google Scholar] [CrossRef] [PubMed]
- Beassoni, P.R.; Gallarato, L.A.; Boetsch, C.; Garrido, M.N.; Lisa, A.T. Pseudomonas aeruginosa exopolyphosphatase is also a polyphosphate: ADP phosphotransferase. Enzym. Res. 2015, 2015, 404607. [Google Scholar] [CrossRef]
- Santos-Torres, M.; Romero-Perdomo, F.; Mendoza-Labrador, J.; Gutiérrez, A.Y.; Vargas, C.; Castro-Rincon, E.; Caro-Quintero, A.; Uribe-Velez, D.; Estrada-Bonilla, G.A. Genomic and phenotypic analysis of rock phosphate-solubilizing rhizobacteria. Rhizosphere 2021, 17, 100290. [Google Scholar] [CrossRef]
- Li, M.; Hao, Y.; Yan, Z.; Kang, E.; Wang, J.; Zhang, K.; Li, Y.; Wu, H.; Kang, X. Long-term degradation from marshes into meadows shifts microbial functional diversity of soil phosphorus cycling in an alpine wetland of the Tibetan Plateau. Land Degrad. Dev. 2021, 33, 628–637. [Google Scholar] [CrossRef]
- Zhong, Z.; Wang, X.; Zhang, X.; Zhang, W.; Xu, Y.; Ren, C.; Han, X.; Yang, G. Edaphic factors but not plant characteristics mainly alter soil microbial properties along a restoration chronosequence of Pinus tabulaeformis stands on Mt. Ziwuling, China. For. Ecol. Manag. 2019, 453, 117625. [Google Scholar] [CrossRef]
- Zhou, Z.Y.; Yu, M.H.; Din, G.D.; Gao, G.L.; He, Y.Y.; Wang, G.Z. Effects of Hedysarum leguminous plants on soil bacterial communities in the Mu Us Desert, northwest China. Ecol. Evol. 2020, 10, 11423–11439. [Google Scholar] [CrossRef] [PubMed]










| P Utilization Level | Plant Species | PPEI | Plant Nutrients (%) | ||
|---|---|---|---|---|---|
| N | P | N:P | |||
| High | ND | 0.076 ± 0.026 | 3.70 ± 0.04 | 0.28 ± 0.01 | 13.22 ± 0.44 |
| MB | 0.063 ± 0.003 | 1.26 ± 0.06 | 0.08 ± 0.01 | 15.79 ± 0.73 | |
| PF | 0.059 ± 0.003 | 1.35 ± 0.08 | 0.08 ± 0.01 | 16.91 ± 0.79 | |
| Average | 0.066 ± 0.008 | 2.10 ± 1.20 | 0.15 ± 0.10 | 15.31 ± 1.74 | |
| Low | TS | 0.036 ± 0.002 | 2.74 ± 0.09 | 0.10 ± 0.004 | 27.43 ± 1.46 |
| AK | 0.032 ± 0.002 | 3.72 ± 0.08 | 0.12 ± 0.01 | 31.12 ± 2.09 | |
| MR | 0.025 ± 0.002 | 1.61 ± 0.08 | 0.04 ± 0.004 | 40.46 ± 3.23 | |
| Average | 0.031 ± 0.005 | 2.69 ± 0.92 | 0.09 ± 0.04 | 33.00 ± 6.17 | |
| Indexes | Plant | Soil | |||||||
|---|---|---|---|---|---|---|---|---|---|
| N | P | N:P | PPEI | Total N | Total P | AN | AP | ||
| Plant | N | 1 | |||||||
| P | 0.728 ** | 1 | |||||||
| N:P | 0.003 | −0.556 * | 1 | ||||||
| PPEI | 0.004 | 0.642 ** | −0.960 ** | 1 | |||||
| Soil | Total N | 0.151 | 0.193 | 0.064 | −0.053 | 1 | |||
| Total P | 0.132 | −0.153 | 0.329 | −0.407 | 0.777 ** | 1 | |||
| AN | 0.030 | 0.207 | −0.095 | 0.103 | 0.968 ** | 0.661 ** | 1 | ||
| AP | 0.316 | 0.527 * | −0.204 | 0.251 | 0.897 ** | 0.474 * | 0.926 ** | 1 | |
| Gene Name | R2 | p-Value | |
|---|---|---|---|
| Phosphomonoesterase | G6PD | 0.265 | <0.05 |
| gpsA | 0.179 | 0.080 | |
| gld | 0.000 | 0.096 | |
| phoA | 0.006 | 0.752 | |
| E3.1.3.8 | 0.158 | 0.102 | |
| suhB | 0.379 | <0.01 | |
| appA | 0.038 | 0.438 | |
| pgpA | 0.005 | 0.771 | |
| phoD | 0.453 | <0.01 | |
| aphA | 0.013 | 0.655 | |
| phoX | 0.080 | 0.254 | |
| phoN | 0.003 | 0.842 | |
| Organic pyrophosphatase | nudF | 0.103 | 0.193 |
| ppx | 0.450 | <0.01 | |
| Gene Name | R2 | p-Value | |
|---|---|---|---|
| Phosphate transport system | pstS | 0.232 | <0.05 |
| pstA | 0.354 | <0.01 | |
| pstB | 0.285 | <0.05 | |
| pstC | 0.305 | <0.05 | |
| pit | 0.025 | 0.527 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, C.; Chen, D.; Zang, L.; Zhang, G.; Liu, Q.; Sui, M.; He, Y.; Wang, S.; Dai, Y.; Wang, L.; et al. The Rhizosphere Functional Microbial Community: A Key Driver of Phosphorus Utilization Efficiency in Karst Forest Plants. Forests 2024, 15, 453. https://doi.org/10.3390/f15030453
Zhou C, Chen D, Zang L, Zhang G, Liu Q, Sui M, He Y, Wang S, Dai Y, Wang L, et al. The Rhizosphere Functional Microbial Community: A Key Driver of Phosphorus Utilization Efficiency in Karst Forest Plants. Forests. 2024; 15(3):453. https://doi.org/10.3390/f15030453
Chicago/Turabian StyleZhou, Chunjie, Danmei Chen, Lipeng Zang, Guangqi Zhang, Qingfu Liu, Mingzhen Sui, Yuejun He, Shasha Wang, Yu Dai, Lidong Wang, and et al. 2024. "The Rhizosphere Functional Microbial Community: A Key Driver of Phosphorus Utilization Efficiency in Karst Forest Plants" Forests 15, no. 3: 453. https://doi.org/10.3390/f15030453