Recent Developments in Microfluidics
A topical collection in Biosensors (ISSN 2079-6374).
Viewed by 60708Editors
Interests: microrobotic sensors; single-cell sensing; bio-MEMS; microfluidics; lab-on-a-chip; organ-on-a-chip; point-of-care biosensors; disease diagnostics; global health
Topical Collection Information
Dear Colleagues,
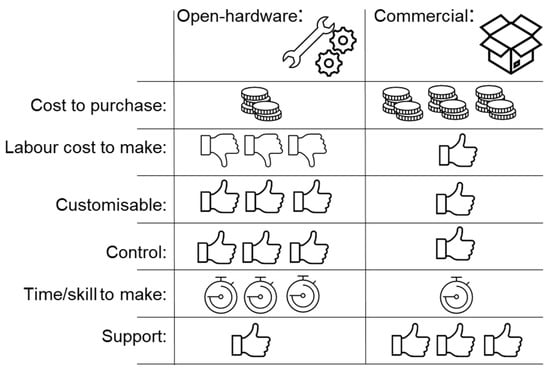
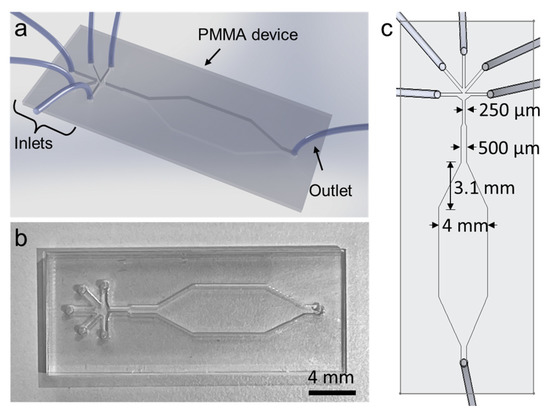
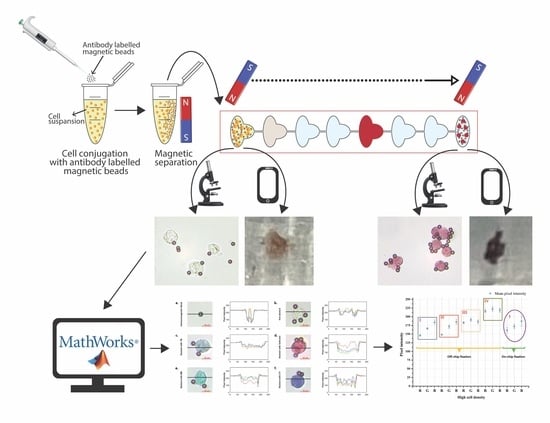
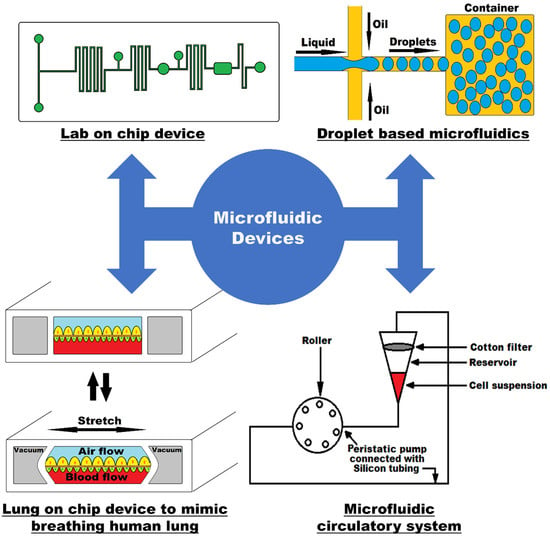
The advent of microfluidics has revolutionized many fields within life sciences by enabling the rapid isolation and detection of molecules and proteins, extracellular vesicles, bacteria, and cells in complex biological fluids with major applications in medical diagnostics, patient monitoring, drug screening, and food safety. Microfluidic technologies are rapidly evolving with new fabrication techniques, channel architecture, and materials (flexible elastomers, paper, 2D materials), along with developments in sensing modalities (optical, electrical, and magnetic) and integrated molecular biology techniques (CRISPR-based approaches). These developments enable faster and higher detection sensitivity in complex environments and challenging applications ranging from unamplified nucleic acid and extracellular vesicle detection to real-time monitoring of physiological signals from wearables. On the other hand, cost-effective, disposable, and simple-use microfluidic technologies are at the frontlines of point-of-care diagnostics and screening of infectious diseases and genetic disorders, especially in low-resource settings; the importance and urgency of which has become evident once more during the recent COVID-19 pandemic.
In this Topical Collection, we are pleased to invite contributions on “Recent Developments in Microfluidics” dedicated to covering the most recent innovations in next-generation microfluidic technologies. Research articles and comprehensive review articles reporting on the latest developments in new fabrication techniques and materials, novel sensing approaches, and applications in molecular and cellular diagnostics and wearable physiological monitoring technologies are of great interest.
Dr. Yunus Alapan
Dr. Yuncheng Man
Collection Editors
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Biosensors is an international peer-reviewed open access monthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript. The Article Processing Charge (APC) for publication in this open access journal is 2700 CHF (Swiss Francs). Submitted papers should be well formatted and use good English. Authors may use MDPI's English editing service prior to publication or during author revisions.
Keywords
- microfluidic biosensors
- lab-on-a-chip
- paper-based microfluidics
- wearable microfluidics
- Microfluidic assays
- molecular diagnostics
- cellular diagnostics
- point-of-care diagnostics
- physiological monitoring
- global health