Machine Learning Approach to Identify Case-Control Studies on ApoE Gene Mutations Linked to Alzheimer’s Disease in Italy
Abstract
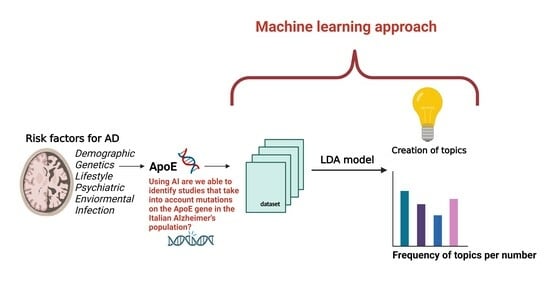
:1. Introduction
2. Materials and Methods
2.1. Paper Location and Selection
2.2. Paper Analysis
- k sets of relevant keywords (each set representing a topic).
- The document-term matrix depicts the statistical relationship between each paper and a specific topic (namely, the topic proportion).
2.3. Results Presentation
3. Results
3.1. Topic 1: Gene Polymorphisms Risk Factor for Alzheimer’s Disease
3.2. Topic 2: The Enigma of Alzheimer’s: Investigating Genetic Patterns, Genotypes as a Future Biomarker
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Blennow, K.; de Leon, M.J.; Zetterberg, H. Alzheimer’s disease. Lancet 2006, 368, 387–403. [Google Scholar] [CrossRef]
- Karch, C.M.; Goate, A.M. Alzheimer’s Disease Risk Genes and Mechanisms of Disease Pathogenesis. Biol. Psychiatry 2015, 77, 43–51. [Google Scholar] [CrossRef]
- Carmona, S.; Hardy, J.; Guerreiro, R. The genetic landscape of Alzheimer disease. Handb. Clin. Neurol. 2018, 148, 395–408. [Google Scholar] [CrossRef]
- Liu, C.-C.; Kanekiyo, T.; Xu, H.; Bu, G. Apolipoprotein E and Alzheimer disease: Risk, mechanisms and therapy. Nat. Rev. Neurol. 2013, 9, 106–118. [Google Scholar] [CrossRef]
- Farrer, L.A.; Cupples, L.A.; Haines, J.L.; Hyman, B.; Kukull, W.A.; Mayeux, R.; Myers, R.H.; Pericak-Vance, M.A.; Risch, N.; van Duijn, C.M. Effects of Age, Sex, and Ethnicity on the Association Between Apolipoprotein E Genotype and Alzheimer Disease. JAMA 1997, 278, 1349–1356. [Google Scholar] [CrossRef]
- Berlau, D.J.; Corrada, M.M.; Head, E.; Kawas, C.H. APOE ε2 is associated with intact cognition but increased Alzheimer pathology in the oldest old. Neurology 2009, 72, 829–834. [Google Scholar] [CrossRef]
- Artiga, M.J.; Bullido, M.J.; Frank, A.; Sastre, I.; Recuero, M.; Garcia, M.A.; Lendon, C.L.; Han, S.W.; Morris, J.C.; Vazquez, J.; et al. Risk for Alzheimer’s disease correlates with transcriptional activity of the APOE gene. Hum. Mol. Genet. 1998, 7, 1887–1892. [Google Scholar] [CrossRef] [PubMed]
- Bekris, L.M.; Millard, S.; Lutz, F.; Li, G.; Galasko, D.R.; Farlow, M.R.; Quinn, J.F.; Kaye, J.A.; Leverenz, J.B.; Tsuang, D.W.; et al. Tau phosphorylation pathway genes and cerebrospinal fluid tau levels in Alzheimer’s disease. Am. J. Med Genet. Part B Neuropsychiatr. Genet. 2012, 159, 874–883. [Google Scholar] [CrossRef] [PubMed]
- Laws, S.M.; Hone, E.; Gandy, S.; Martins, R.N. Expanding the association between the APOE gene and the risk of Alzheimer’s disease: Possible roles for APOE promoter polymorphisms and alterations in APOE transcription. J. Neurochem. 2003, 84, 1215–1236. [Google Scholar] [CrossRef] [PubMed]
- Roses, A.D. A model for susceptibility polymorphisms for complex diseases: Apolipoprotein E and Alzheimer disease. Neurogenetics 1997, 1, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Yuryevna Bokovnya, A.; Rustamovich Begishev, I.; Ilduzovna Khisamova, Z.; Rashidovna Narimanova, N.; Mikhailovna Sherbakova, L. Legal Approaches to Artificial Intelligence Concept and Essence Definition. Rev. San Gregor. 2020, 1, 115–121. [Google Scholar] [CrossRef]
- Shapiro, S.C. Artificial intelligence. In Encyclopedia of Artificial Intelligence, 2nd ed.; John Wiley & Sons: New York, NY, USA, 1992; Volume 1. [Google Scholar]
- Mitchell, T.M. Machine Learning; Mcgraw-Hill International: New York, NY, USA, 1997. [Google Scholar]
- Demner-Fushman, D.; Lin, J. Answering Clinical Questions with Knowledge-Based and Statistical Techniques. Comput. Linguist. 2007, 33, 63–103. [Google Scholar] [CrossRef]
- Jones, B.D.; Baumgartner, F.R. The Politics of Attention: How Government Prioritizes Problems; University of Chicago Press: Chichago, IL, USA, 2005. [Google Scholar]
- King, G.; Lowe, W. An Automated Information Extraction Tool for International Conflict Data with Performance as Good as Human Coders: A Rare Events Evaluation Design. Int. Organ. 2003, 57, 617–642. [Google Scholar] [CrossRef]
- Topol, E.J. High-performance medicine: The convergence of human and artificial intelligence. Nat. Med. 2019, 25, 44–56. [Google Scholar] [CrossRef]
- Hashimoto, D.A.; Rosman, G.; Rus, D.; Meireles, O.R.M. Artificial Intelligence in Surgery: Promises and Perils. Ann. Surg. 2018, 268, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Bettencourt, C.; Skene, N.; Bandres-Ciga, S.; Anderson, E.; Winchester, L.M.; Foote, I.F.; Schwartzentruber, J.; Botia, J.A.; Nalls, M.; Singleton, A.; et al. Artificial intelligence for dementia genetics and omics. Alzheimer’s Dement. 2023, 19, 5905–5921. [Google Scholar] [CrossRef] [PubMed]
- Blei, D.M.; Ng, A.Y.; Jordan, M.I. Latent Dirichlet Allocation. J. Mach. Learn. Res. 2003, 3, 993–1022. [Google Scholar]
- Ammirato, S.; Felicetti, A.M.; Rogano, D.; Linzalone, R.; Corvello, V. Digitalising the Systematic Literature Review process: The MySLR platform. Knowl. Manag. Res. Pract. 2022, 21, 777–794. [Google Scholar] [CrossRef]
- Denyer, D.; Tranfield, D. Producing a systematic review. In The Sage Handbook of Organizational Research Methods; Sage Publications Ltd.: Thousand Oaks, CA, USA, 2009; pp. 671–689. [Google Scholar]
- Chen, Z.; Liu, B. Topic Modeling using Topics from Many Domains, Lifelong Learning and Big Data. Proc. Mach. Learn. Res. 2014, 32, 703–711. Available online: https://proceedings.mlr.press/v32/chenf14.html (accessed on 20 February 2024).
- Yi, X.; Allan, J. Evaluating topic models for information retrieval. In Proceedings of the 17th ACM Conference on Information and Knowledge Management, Napa Valley, CA, USA, 26–30 October 2008; Association for Computing Machinery: New York, NY, USA, 2008; pp. 1431–1432. [Google Scholar]
- Ballard, C.; Gauthier, S.; Corbett, A.; Brayne, C.; Aarsland, D.; Jones, E. Alzheimer’s disease. Lancet 2011, 377, 1019–1031. [Google Scholar] [CrossRef] [PubMed]
- Bertram, L.; Tanzi, R.E. Genome-wide association studies in Alzheimer’s disease. Hum. Mol. Genet. 2009, 18, R137–R145. [Google Scholar] [CrossRef] [PubMed]
- Albani, D.; Tettamanti, M.; Batelli, S.; Polito, L.; Dusi, S.; Ateri, E.; Forloni, G.; Lucca, U. Interleukin-1α, interleukin-1β and tumor necrosis factor-α genetic variants and risk of dementia in the very old: Evidence from the “Monzino 80-plus” prospective study. AGE 2012, 34, 519–526. [Google Scholar] [CrossRef]
- Andreoli, V.; De Marco, E.V.; Trecroci, F.; Cittadella, R.; Di Palma, G.; Gambardella, A. Potential involvement of GRIN2B encoding the NMDA receptor subunit NR2B in the spectrum of Alzheimer’s disease. J. Neural Transm. 2014, 121, 533–542. [Google Scholar] [CrossRef]
- Bagnoli, S.; Cellini, E.; Tedde, A.; Nacmias, B.; Piacentini, S.; Bessi, V.; Bracco, L.; Sorbi, S. Association of IL10 promoter polymorphism in Italian Alzheimer’s disease. Neurosci. Lett. 2007, 418, 262–265. [Google Scholar] [CrossRef] [PubMed]
- Bagnoli, S.; Piaceri, I.; Tedde, A.; Bessi, V.; Bracco, L.; Sorbi, S.; Nacmias, B. Tomm40 polymorphisms in Italian Alzheimer’s disease and frontotemporal dementia patients. Neurol. Sci. 2013, 34, 995–998. [Google Scholar] [CrossRef] [PubMed]
- Bartoletti-Stella, A.; Tarozzi, M.; Mengozzi, G.; Asirelli, F.; Brancaleoni, L.; Mometto, N.; Stanzani-Maserati, M.; Baiardi, S.; Linarello, S.; Spallazzi, M.; et al. Dementia-related genetic variants in an Italian population of early-onset Alzheimer’s disease. Front. Aging Neurosci. 2022, 14, 969817. [Google Scholar] [CrossRef]
- Belloy, M.E.; Eger, S.J.; Le Guen, Y.; Damotte, V.; Ahmad, S.; Ikram, M.A.; Ramirez, A.; Tsolaki, A.C.; Rossi, G.; Jansen, I.E.; et al. Challenges at the APOE locus: A robust quality control approach for accurate APOE genotyping. Alzheimer’s Res. Ther. 2022, 14, 22. [Google Scholar] [CrossRef]
- Bizzarro, A.; Seripa, D.; Acciarri, A.; Matera, M.G.; Pilotto, A.; Tiziano, F.D.; Brahe, C.; Masullo, C. The complex interaction between APOE promoter and AD: An Italian case–control study. Eur. J. Hum. Genet. 2009, 17, 938–945. [Google Scholar] [CrossRef]
- Bosco, P.; Ferri, R.; Salluzzo, M.G.; Castellano, S.; Signorelli, M.; Nicoletti, F.; Di Nuovo, S.; Drago, F.; Caraci, F. Role of the Transforming-Growth-Factor-β1 Gene in Late-Onset Alzheimer’s Disease: Implications for the Treatment. Curr. Genom. 2013, 14, 147–156. [Google Scholar] [CrossRef]
- Broer, L.; Buchman, A.S.; Deelen, J.; Evans, D.S.; Faul, J.D.; Lunetta, K.L.; Sebastiani, P.; Smith, J.A.; Smith, A.V.; Tanaka, T.; et al. GWAS of longevity in CHARGE consortium confirms APOE and FOXO3 candidacy. J. Gerontol. Ser. A Biomed. Sci. Med. Sci. 2015, 70, 110–118. [Google Scholar] [CrossRef]
- Bucossi, S.; Polimanti, R.; Mariani, S.; Ventriglia, M.; Bonvicini, C.; Migliore, S.; Manfellotto, D.; Salustri, C.; Vernieri, F.; Rossini, P.M.; et al. Association of K832R and R952K SNPs of Wilson’s disease gene with Alzheimer’s disease. J. Alzheimer’s Dis. 2012, 29, 913–919. [Google Scholar] [CrossRef]
- Capurso, C.; Solfrizzi, V.; Colacicco, A.M.; D’Introno, A.; Frisardi, V.; Imbimbo, B.P.; Lorusso, M.; Vendemiale, G.; Denitto, M.; Santamato, A.; et al. Interleukin 6–174 G/C promoter and variable number of tandem repeats (VNTR) gene polymorphisms in sporadic Alzheimer’s disease. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2010, 34, 177–182. [Google Scholar] [CrossRef]
- Capurso, C.; Panza, F.; Seripa, D.; Frisardi, V.; Imbimbo, B.P.; Verdile, G.; Vendemiale, G.; Pilotto, A.; Solfrizzi, V. Polymorphisms in Glutathione S-Transferase Omega-1 Gene and Increased Risk of Sporadic Alzheimer Disease. Rejuvenation Res. 2010, 13, 645–652. [Google Scholar] [CrossRef]
- Cellini, E.; Tedde, A.; Bagnoli, S.; Pradella, S.; Piacentini, S.; Sorbi, S.; Nacmias, B. Implication of Sex and SORL1 Variants in Italian Patients with Alzheimer Disease. Arch. Neurol. 2009, 66, 1260–1266. [Google Scholar] [CrossRef] [PubMed]
- Ciminelli, B.M.; Menduti, G.; Benussi, L.; Ghidoni, R.; Binetti, G.; Squitti, R.; Rongioletti, M.; Nica, S.; Novelletto, A.; Rossi, L.; et al. Polymorphic Genetic Markers of the GABA Catabolism Pathway in Alzheimer’s Disease. J. Alzheimer’s Dis. 2020, 77, 301–311. [Google Scholar] [CrossRef] [PubMed]
- Clarelli, F.; Mascia, E.; Santangelo, R.; Mazzeo, S.; Giacalone, G.; Galimberti, D.; Fusco, F.; Zuffi, M.; Fenoglio, C.; Franceschi, M.; et al. CHRNA7 Gene and Response to Cholinesterase Inhibitors in an Italian Cohort of Alzheimer’s Disease Patients. J. Alzheimer’s Dis. 2016, 52, 1203–1208. [Google Scholar] [CrossRef] [PubMed]
- Colacicco, A.M.; Solfrizzi, V.; D’introno, A.; Capurso, C.; Kehoe, P.G.; Seripa, D.; Pilotto, A.; Santamato, A.; Capurso, A.; Panza, F. Alpha-2-macroglobulin gene, oxidized low-density lipoprotein receptor-1 locus, and sporadic Alzheimer’s disease. Neurobiol. Aging 2009, 30, 1518–1520. [Google Scholar] [CrossRef] [PubMed]
- Corbo, R.M.; Gambina, G.; Ulizzi, L.; Moretto, G.; Scacchi, R. Genetic Variation of CYP19 (Aromatase) Gene Influences Age at Onset of Alzheimer’s Disease in Women. Dement. Geriatr. Cogn. Disord. 2009, 27, 513–518. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Sanabria, F.; Bonilla-Vargas, K.; Estrada, K.; Mancera, O.; Vega, E.; Guerrero, E.; Ortega-Rojas, J.; María, F.M.; Romero, A.; Montañés, P.; et al. Analysis of cognitive performance and polymorphisms of SORL1, PVRL2, CR1, TOMM40, APOE, PICALM, GWAS_14q, CLU, and BIN1 in patients with mild cognitive impairment and cognitively healthy controls. Neurologia 2021, 36, 681–691. [Google Scholar] [CrossRef] [PubMed]
- DeMichele-Sweet, M.A.A.; Klei, L.; Creese, B.; Harwood, J.C.; Weamer, E.A.; McClain, L.; Sims, R.; Hernandez, I.; Moreno-Grau, S.; Tárraga, L.; et al. Genome-wide association identifies the first risk loci for psychosis in Alzheimer disease. Mol. Psychiatry 2021, 26, 5797–5811. [Google Scholar] [CrossRef] [PubMed]
- Di Maria, E.; Giorgio, E.; Uliana, V.; Bonvicini, C.; Faravelli, F.; Cammarata, S.; Novello, M.C.; Galimberti, D.; Scarpini, E.; Zanetti, O.; et al. Possible Influence of a Non-Synonymous Polymorphism Located in the NGF Precursor on Susceptibility to Late-Onset Alzheimer’s Disease and Mild Cognitive Impairment. J. Alzheimer’s Dis. 2012, 29, 699–705. [Google Scholar] [CrossRef]
- Emanuele, E.; Lista, S.; Ghidoni, R.; Binetti, G.; Cereda, C.; Benussi, L.; Maletta, R.; Bruni, A.C.; Politi, P. Chromosome 9p21.3 genotype is associated with vascular dementia and Alzheimer’s disease. Neurobiol. Aging 2011, 32, 1231–1235. [Google Scholar] [CrossRef]
- Finckh, U.; van Hadeln, K.; Müller-Thomsen, T.; Alberici, A.; Binetti, G.; Hock, C.; Nitsch, R.M.; Stoppe, G.; Reiss, J.; Gal, A. Association of late-onset Alzheimer disease with a genotype of PLAU, the gene encoding urokinase-type plasminogen activator on chromosome 10q22.2. Neurogenetics 2003, 4, 213–217. [Google Scholar] [CrossRef]
- Fortney, K.; Dobriban, E.; Garagnani, P.; Pirazzini, C.; Monti, D.; Mari, D.; Atzmon, G.; Barzilai, N.; Franceschi, C.; Owen, A.B.; et al. Genome-Wide Scan Informed by Age-Related Disease Identifies Loci for Exceptional Human Longevity. PLoS Genet. 2015, 11, e1005728. [Google Scholar] [CrossRef] [PubMed]
- Galimberti, D.; Scarpini, E.; Venturelli, E.; Strobel, A.; Herterich, S.; Fenoglio, C.; Guidi, I.; Scalabrini, D.; Cortini, F.; Bresolin, N.; et al. Association of a NOS1 promoter repeat with Alzheimer’s disease. Neurobiol. Aging 2008, 29, 1359–1365. [Google Scholar] [CrossRef]
- Guerini, F.R.; Farina, E.; Costa, A.S.; Baglio, F.; Saibene, F.L.; Margaritella, N.; Calabrese, E.; Zanzottera, M.; Bolognesi, E.; Nemni, R.; et al. ApoE and SNAP-25 Polymorphisms Predict the Outcome of Multidimensional Stimulation Therapy Rehabilitation in Alzheimer’s Disease. Neurorehabilit. Neural Repair 2016, 30, 883–893. [Google Scholar] [CrossRef] [PubMed]
- Hollingworth, P.; Harold, D.; Sims, R.; Gerrish, A.; Lambert, J.-C.; Carrasquillo, M.M.; Abraham, R.; Hamshere, M.L.; Pahwa, J.S.; Moskvina, V.; et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat. Genet. 2011, 43, 429–435. [Google Scholar] [CrossRef] [PubMed]
- Jun, G.; Ibrahim-Verbaas, C.A.; Vronskaya, M.; Lambert, J.C.; Chung, J.; Naj, A.C.; Kunkle, B.W.; Wang, L.S.; Bis, J.C.; Bel-lenguez, C.; et al. A novel Alzheimer disease locus located near the gene encoding tau protein. Mol. Psychiatry 2016, 21, 108–117. [Google Scholar] [CrossRef]
- Lambert, J.-C.; Heath, S.; Even, G.; Campion, D.; Sleegers, K.; Hiltunen, M.; Combarros, O.; Zelenika, D.; Bullido, M.J.; Tavernier, B.; et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat. Genet. 2009, 41, 1094–1099. [Google Scholar] [CrossRef]
- Lambert, J.C.; Ibrahim-Verbaas, C.A.; Harold, D.; Naj, A.C.; Sims, R.; Bellenguez, C.; DeStafano, A.L.; Bis, J.C.; Beecham, G.W.; Grenier-Boley, B.; et al. Meta-Analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet. 2013, 45, 1452–1458. [Google Scholar] [CrossRef]
- Lanni, C.; Garbin, G.; Lisa, A.; Biundo, F.; Ranzenigo, A.; Sinforiani, E.; Cuzzoni, G.; Govoni, S.; Ranzani, G.N.; Racchi, M. Influence of COMT Val158Met Polymorphism on Alzheimer’s Disease and Mild Cognitive Impairment in Italian Patients. J. Alzheimer’s Dis. 2012, 32, 919–926. [Google Scholar] [CrossRef]
- Laws, S.M.; Perneczky, R.; Wagenpfeil, S.; Müller, U.; Förstl, H.; Martins, R.N.; Kurz, A.; Riemenschneider, M. TNF polymorphisms in Alzheimer disease and functional implications on CSF beta-amyloid levels. Hum. Mutat. 2005, 26, 29–35. [Google Scholar] [CrossRef]
- Lescai, F.; Pirazzini, C.; D’Agostino, G.; Santoro, A.; Ghidoni, R.; Benussi, L.; Galimberti, D.; Federica, E.; Marchegiani, F.; Cardelli, M.; et al. Failure to Replicate an Association of rs5984894 SNP in the PCDH11X Gene in a Collection of 1222 Alzheimer’s Disease Affected Patients. J. Alzheimer’s Dis. 2010, 21, 385–388. [Google Scholar] [CrossRef] [PubMed]
- Lescai, F.; Chiamenti, A.M.; Codemo, A.; Pirazzini, C.; D’Agostino, G.; Ruaro, C.; Ghidoni, R.; Benussi, L.; Galimberti, D.; Esposito, F.; et al. An APOE Haplotype Associated with Decreased ε4 Expression Increases the Risk of Late Onset Alzheimer’s Disease. J. Alzheimer’s Dis. 2011, 24, 235–245. [Google Scholar] [CrossRef] [PubMed]
- Licastro, F.; Chiappelli, M.; Caldarera, C.M.; Porcellini, E.; Carbone, I.; Caruso, C.; Lio, D.; Corder, E.H. Sharing Pathogenetic Mechanisms between Acute Myocardial Infarction and Alzheimer’s Disease as Shown by Partially Overlapping of Gene Variant Profiles. J. Alzheimer’s Dis. 2011, 23, 421–431. [Google Scholar] [CrossRef]
- Licastro, F.; Raschi, E.; Carbone, I.; Porcellini, E. Variants in Antiviral Genes are Risk Factors for Cognitive Decline and Dementia. J. Alzheimer’s Dis. 2015, 46, 655–663. [Google Scholar] [CrossRef]
- Lio, D.; Licastro, F.; Scola, L.; Chiappelli, M.; Grimaldi, L.M.; Crivello, A.; Colonna-Romano, G.; Candore, G.; Franceschi, C.; Caruso, C. Interleukin-10 promoter polymorphism in sporadic Alzheimer’s disease. Genes Immun. 2003, 4, 234–238. [Google Scholar] [CrossRef]
- Lio, D.; Annoni, G.; Licastro, F.; Crivello, A.; Forte, G.I.; Scola, L.; Colonna-Romano, G.; Candore, G.; Arosio, B.; Galimberti, L.; et al. Tumor necrosis factor-α−308A/G polymorphism is associated with age at onset of Alzheimer’s disease. Mech. Ageing Dev. 2006, 127, 567–571. [Google Scholar] [CrossRef]
- Lu, R.-C.; Yang, W.; Tan, L.; Sun, F.-R.; Tan, M.-S.; Zhang, W.; Wang, H.-F.; Tan, L. Association of HLA-DRB1 polymorphism with Alzheimer’s disease: A replication and meta-analysis. Oncotarget 2017, 8, 93219–93226. [Google Scholar] [CrossRef] [PubMed]
- Lupton, M.K.; Strike, L.; Hansell, N.K.; Wen, W.; Mather, K.A.; Armstrong, N.J.; Thalamuthu, A.; McMahon, K.L.; de Zubicaray, G.I.; Assareh, A.A.; et al. The effect of increased genetic risk for Alzheimer’s disease on hippocampal and amygdala volume. Neurobiol. Aging 2016, 40, 68–77. [Google Scholar] [CrossRef]
- Maletta, R.; Smirne, N.; Bernardi, L.; Anfossi, M.; Gallo, M.; Conidi, M.E.; Colao, R.; Puccio, G.; Curcio, S.A.; Laganà, V.; et al. Frequency of Cardiovascular Genetic Risk Factors in a Calabrian Population and Their Effects on Dementia. J. Alzheimer’s Dis. 2018, 61, 1179–1187. [Google Scholar] [CrossRef] [PubMed]
- Mariani, S.; Ventriglia, M.; Simonelli, I.; Spalletta, G.; Bucossi, S.; Siotto, M.; Assogna, F.; Melgari, J.M.; Vernieri, F.; Squitti, R. Effects of hemochromatosis and transferrin gene mutations on peripheral iron dyshomeostasis in mild cognitive impairment and Alzheimer’s and Parkinson’s diseases. Front. Aging Neurosci. 2013, 5, 37. [Google Scholar] [CrossRef] [PubMed]
- Masri, I.; Salami, A.; El Shamieh, S.; Bissar-Tadmouri, N. rs3851179G>A in PICALM is Protective Against Alzheimer’s Disease in Five Different Countries Surrounding the Mediterranean. Curr. Aging Sci. 2020, 13, 162–168. [Google Scholar] [CrossRef] [PubMed]
- Minoretti, P.; Gazzaruso, C.; Di Vito, C.; Emanuele, E.; Bianchi, M.; Coen, E.; Reino, M.; Geroldi, D. Effect of the functional toll-like receptor 4 Asp299Gly polymorphism on susceptibility to late-onset Alzheimer’s disease. Neurosci. Lett. 2006, 391, 147–149. [Google Scholar] [CrossRef]
- Montesanto, A.; Crocco, P.; Anfossi, M.; Smirne, N.; Puccio, G.; Colao, R.; Maletta, R.; Passarino, G.; Bruni, A.C.; Rose, G. The Genetic Variability of UCP4 Affects the Individual Susceptibility to Late-Onset Alzheimer’s Disease and Modifies the Disease’s Risk in APOE-ɛ4 Carriers. J. Alzheimer’s Dis. 2016, 51, 1265–1274. [Google Scholar] [CrossRef] [PubMed]
- Nacmias, B.; Tedde, A.; Bagnoli, S.; Cellini, E.; Guarnieri, B.M.; Piacentini, S.; Sorbi, S. Implication of GAB2 Gene Polymorphism in Italian Patients with Alzheimer’s Disease. J. Alzheimer’s Dis. 2009, 16, 513–515. [Google Scholar] [CrossRef]
- Napolioni, V.; Giannì, P.; Carpi, F.M.; Predazzi, I.M.; Lucarini, N. APOE haplotypes are associated with human longevity in a Central Italy population: Evidence for epistasis with HP 1/2 polymorphism. Clin. Chim. Acta 2011, 412, 1821–1824. [Google Scholar] [CrossRef] [PubMed]
- Olgiati, P.; Politis, A.; Albani, D.; Rodilossi, S.; Polito, L.; Zisaki, A.; Piperi, C.; Liappas, I.; Stamouli, E.; Mailis, A.; et al. Effects of SORL1 gene on Alzheimer’s disease. Focus on gender, neuropsychiatric symptoms and pro-inflammatory cytokines. Curr. Alzheimer Res. 2013, 10, 154–164. [Google Scholar] [CrossRef]
- Orlacchio, A.; Kawarai, T.; Polidoro, M.; Stefani, A.; Orlacchio, A.; George-Hyslop, P.H.S.; Bernardi, G. Association analysis between Alzheimer’s disease and the Nicastrin gene polymorphisms. Neurosci. Lett. 2002, 333, 115–118. [Google Scholar] [CrossRef]
- Pilotto, A.; Franceschi, M.; D’Onofrio, G.; Bizzarro, A.; Mangialasche, F.; Cascavilla, L.; Paris, F.; Matera, M.G.; Pilotto, A.; Daniele, A.; et al. Effect of a CYP2D6 polymorphism on the efficacy of donepezil in patients with Alzheimer disease. Neurology 2009, 73, 761–767. [Google Scholar] [CrossRef]
- Pola, R.; Flex, A.; Gaetani, E.; Proia, A.S.; Papaleo, P.; Di Giorgio, A.; Straface, G.; Pecorini, G.; Serricchio, M.; Pola, P. Monocyte chemoattractant protein-1 (MCP-1) gene polymorphism and risk of Alzheimer’s disease in Italians. Exp. Gerontol. 2004, 39, 1249–1252. [Google Scholar] [CrossRef]
- Poleggi, A.; Bizzarro, A.; Acciarri, A.; Antuono, P.; Bagnoli, S.; Cellini, E.; Forno, G.D.; Giannattasio, C.; Lauria, A.; Matera, M.G.; et al. Codon 129 polymorphism of prion protein gene in sporadic Alzheimer’s disease. Eur. J. Neurol. 2008, 15, 173–178. [Google Scholar] [CrossRef]
- Poli, M.; Gatta, L.B.; Lovati, C.; Mariani, C.; Galimberti, D.; Scarpini, E.; Biunno, I.; Musicco, M.; Dominici, R.; Albertini, A.; et al. Interaction between the APOE ɛ4 allele and the APH-1b c+651T>G SNP in Alzheimer’s disease. Neurobiol. Aging 2008, 29, 1494–1501. [Google Scholar] [CrossRef]
- Porrello, E.; Monti, M.C.; Sinforiani, E.; Cairati, M.; Guaita, A.; Montomoli, C.; Govoni, S.; Racchi, M. Estrogen receptor α and APOEɛ4 polymorphisms interact to increase risk for sporadic AD in Italian females. Eur. J. Neurol. 2006, 13, 639–644. [Google Scholar] [CrossRef] [PubMed]
- Scacchi, R.; Gambina, G.; Moretto, G.; Corbo, R.M. Association study between P53 and P73 gene polymorphisms and the sporadic late-onset form of Alzheimer’s disease. J. Neural Transm. 2009, 116, 1179–1184. [Google Scholar] [CrossRef]
- Scassellati, C.; Zanardini, R.; Squitti, R.; Bocchio-Chiavetto, L.; Bonvicini, C.; Binetti, G.; Zanetti, O.; Cassetta, E.; Gennarelli, M. Promoter haplotypes of interleukin-10 gene and sporadic Alzheimer’s disease. Neurosci. Lett. 2004, 356, 119–122. [Google Scholar] [CrossRef]
- Schmidt, R.; Schmidt, H.; Haybaeck, J.; Loitfelder, M.; Weis, S.; Cavalieri, M.; Seiler, S.; Enzinger, C.; Ropele, S.; Erkinjuntti, T.; et al. Heterogeneity in age-related white matter changes. Acta Neuropathol. 2011, 122, 171–185. [Google Scholar] [CrossRef]
- Schott, J.M.; Crutch, S.J.; Carrasquillo, M.M.; Uphill, J.; Shakespeare, T.J.; Ryan, N.S.; Yong, K.X.; Lehmann, M.; Boeve, B.F.; Murray, M.E.; et al. Genetic risk factors for the posterior cortical atrophy variant of Alzheimer’s disease. Alzheimer’s Dement. 2016, 12, 862–871. [Google Scholar] [CrossRef]
- Scola, L.; Licastro, F.; Chiappelli, M.; Franceschi, C.; Grimaldi, L.M.; Crivello, A.; Colonna-Romano, G.; Candore, G.; Lio, D.; Caruso, C. Allele frequencies of +874T→A single nucleotide polymorphism at the first intron of IFN-γ gene in Alzheimer’s disease patients. Aging Clin. Exp. Res. 2003, 15, 292–295. [Google Scholar] [CrossRef] [PubMed]
- Seripa, D.; Matera, M.G.; Franceschi, M.; Bizzarro, A.; Paris, F.; Cascavilla, L.; Rinaldi, M.; Panza, F.; Solfrizzi, V.; Daniele, A.; et al. Association Analysis of GRIN2B, Encoding N-Methyl-D-Aspartate Receptor 2B Subunit, and Alzheimer’s Disease. Dement. Geriatr. Cogn. Disord. 2008, 25, 287–292. [Google Scholar] [CrossRef] [PubMed]
- Seripa, D.; Matera, M.G.; Franceschi, M.; Daniele, A.; Bizzarro, A.; Rinaldi, M.; Panza, F.; Fazio, V.M.; Gravina, C.; D’Onofrio, G.; et al. The RELN Locus in Alzheimer’s Disease. J. Alzheimer’s Dis. 2008, 14, 335–344. [Google Scholar] [CrossRef] [PubMed]
- Seripa, D.; D’Onofrio, G.; Panza, F.; Cascavilla, L.; Masullo, C.; Pilotto, A. The Genetics of the Human APOE Polymorphism. Rejuvenation Res. 2011, 14, 491–500. [Google Scholar] [CrossRef]
- Serpente, M.; Fenoglio, C.; Villa, C.; Cortini, F.; Cantoni, C.; Ridolfi, E.; Clerici, F.; Marcone, A.; Benussi, L.; Ghidoni, R.; et al. Role of OLR1 and Its Regulating hsa-miR369-3p in Alzheimer’s Disease: Genetics and Expression Analysis. J. Alzheimer’s Dis. 2011, 26, 787–793. [Google Scholar] [CrossRef] [PubMed]
- Squillario, M.; Abate, G.; Tomasi, F.; Tozzo, V.; Barla, A.; Uberti, D.; Weiner, M.W.; Aisen, P.; Petersen, R.; Clifford, J.R.; et al. A telescope GWAS analysis strategy, based on SNPs-genes-pathways ensamble and on multivariate algorithms, to characterize late onset Alzheimer’s disease. Sci. Rep. 2020, 10, 12063. [Google Scholar] [CrossRef] [PubMed]
- Talwar, P.; Kushwaha, S.; Rawat, C.; Kaur, H.; Srivastava, A.; Agarwal, R.; Chandna, P.; Tucci, P.; Saso, L.; Kukreti, R. Validating a Genomic Convergence and Network Analysis Approach Using Association Analysis of Identified Candidate Genes in Alzheimer’s Disease. Front. Genet. 2021, 12, 722221. [Google Scholar] [CrossRef]
- Tedde, A.; Bagnoli, S.; Piaceri, I.; Lucenteforte, E.; Bessi, V.; Bracco, L.; Mugelli, A.; Sorbi, S.; Nacmias, B. Different implication of NEDD9 genetic variant in early and late-onset Alzheimer’s disease. Neurosci. Lett. 2010, 477, 121–123. [Google Scholar] [CrossRef]
- Tindale, L.C.; Leach, S.; Spinelli, J.J.; Brooks-Wilson, A.R. Lipid and Alzheimer’s disease genes associated with healthy aging and longevity in healthy oldest-old. Oncotarget 2017, 8, 20612–20621. [Google Scholar] [CrossRef]
- Tisato, V.; Zuliani, G.; Vigliano, M.; Longo, G.; Franchini, E.; Secchiero, P.; Zauli, G.; Paraboschi, E.M.; Singh, A.V.; Serino, M.L.; et al. Gene-gene interactions among coding genes of iron-homeostasis proteins and APOE-alleles in cognitive impairment diseases. PLoS ONE 2018, 13, e0193867. [Google Scholar] [CrossRef]
- Valenza, A.; Bizzarro, A.; Marra, C.; Lauria, A.; Guglielmi, V.; Rossi, C.; Tiziano, F.; Brahe, C.; Masullo, C. The APOE-491 A/T promoter polymorphism effect on cognitive profile of Alzheimer’s patients. Neurosci. Lett. 2010, 472, 199–203. [Google Scholar] [CrossRef]
- Venturelli, E.; Galimberti, D.; Lovati, C.; Fenoglio, C.; Scalabrini, D.; Mariani, C.; Forloni, G.; Bresolin, N.; Scarpini, E. The T-786C NOS3 polymorphism in Alzheimer’s disease: Association and influence on gene expression. Neurosci. Lett. 2005, 382, 300–303. [Google Scholar] [CrossRef]
- Wang, Z.; Lei, H.; Zheng, M.; Li, Y.; Cui, Y.; Hao, F. Meta-analysis of the Association between Alzheimer Disease and Variants in GAB2, PICALM, and SORL1. Mol. Neurobiol. 2016, 53, 6501–6510. [Google Scholar] [CrossRef] [PubMed]
- Bernardi, L.; Frangipane, F.; Smirne, N.; Colao, R.; Puccio, G.; Curcio, S.A.; Mirabelli, M.; Maletta, R.; Anfossi, M.; Gallo, M.; et al. Epidemiology and genetics of frontotemporal dementia: A door-to-door survey in Southern Italy. Neurobiol. Aging 2012, 33, 2948.e1–2948.e10. [Google Scholar] [CrossRef] [PubMed]
- Calabretta, A.; Tedeschi, T.; Di Cola, G.; Corradini, R.; Sforza, S.; Marchelli, R. Arginine-based PNA microarrays for APOE genotyping. Mol. Biosyst. 2009, 5, 1323–1330. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.-C.; Sun, Y.; Lai, L.-C.; Chen, S.-Y.; Lee, W.-C.; Chen, J.-H.; Chen, T.-F.; Chen, H.-H.; Wen, L.-L.; Yip, P.-K.; et al. Genetic polymorphisms of nerve growth factor receptor (NGFR) and the risk of Alzheimer’s disease. J. Negat. Results Biomed. 2012, 11, 5. [Google Scholar] [CrossRef] [PubMed]
- de Rojas, I.; Moreno-Grau, S.; Tesi, N.; Grenier-Boley, B.; Andrade, V.; Jansen, I.E.; Pedersen, N.L.; Stringa, N.; Zettergren, A.; Hernández, I.; et al. Common variants in Alzheimer’s disease and risk stratification by polygenic risk scores. Nat. Commun. 2021, 12, 3417. [Google Scholar] [CrossRef] [PubMed]
- Flex, A.; Gaetani, E.; Proia, A.S.; Pecorini, G.; Straface, G.; Biscetti, F.; Fioroni, G.; Sabusco, A.; Flore, R.; Tondi, P.; et al. Analysis of functional polymorphisms of metalloproteinase genes in persons with vascular dementia and Alzheimer’s disease. J. Gerontol. Ser. A 2006, 61, 1065–1069. [Google Scholar] [CrossRef]
- Grünblatt, E.; Reif, A.; Jungwirth, S.; Galimberti, D.; Weber, H.; Scarpini, E.; Sauer, C.; Wichart, I.; Rainer, M.K.; Huber, K.; et al. Genetic variation in the choline O-acetyltransferase gene in depression and Alzheimer’s disease: The VITA and Milano studies. J. Psychiatr. Res. 2011, 45, 1250–1256. [Google Scholar] [CrossRef] [PubMed]
- Hansmannel, F.; Lendon, C.; Pasquier, F.; Dumont, J.; Hannequin, D.; Chapuis, J.; Laumet, G.; Ayral, A.-M.; Galimberti, D.; Scarpini, E.; et al. Is the ornithine transcarbamylase gene a genetic determinant of Alzheimer’s disease? Neurosci. Lett. 2009, 449, 76–80. [Google Scholar] [CrossRef]
- Hong, S.; Prokopenko, D.; Dobricic, V.; Kilpert, F.; Bos, I.; Vos, S.J.B.; Tijms, B.M.; Andreasson, U.; Blennow, K.; Vandenberghe, R.; et al. Genome-wide association study of Alzheimer’s disease CSF biomarkers in the EMIF-AD Multimodal Biomarker Discovery dataset. Transl. Psychiatry 2020, 10, 403. [Google Scholar] [CrossRef]
- Krumbiegel, M.M.; Pasutto, F.; Mardin, C.Y.; Weisschuh, N.; Paoli, D.; Gramer, E.; Weber, B.H.; Kruse, F.E.; Schlötzer-Schrehardt, U.; Reis, A. Apolipoprotein E Genotypes in Pseudoexfoliation Syndrome and Pseudoexfoliation Glaucoma. Eur. J. Gastroenterol. Hepatol. 2010, 19, 561–565. [Google Scholar] [CrossRef]
- Lambert, J.-C.; Zelenika, D.; Hiltunen, M.; Chouraki, V.; Combarros, O.; Bullido, M.J.; Tognoni, G.; Fiévet, N.; Boland, A.; Arosio, B.; et al. Evidence of the association of BIN1 and PICALM with the AD risk in contrasting European populations. Neurobiol. Aging 2011, 32, 756.e11–756.e15. [Google Scholar] [CrossRef] [PubMed]
- Lambert, J.-C.; Grenier-Boley, B.; Harold, D.; Zelenika, D.; Chouraki, V.; Kamatani, Y.; Sleegers, K.; A Ikram, M.; Hiltunen, M.; Reitz, C.; et al. Genome-wide haplotype association study identifies the FRMD4A gene as a risk locus for Alzheimer’s disease. Mol. Psychiatry 2013, 18, 461–470. [Google Scholar] [CrossRef] [PubMed]
- Mazzeo, S.; Bessi, V.; Padiglioni, S.; Bagnoli, S.; Bracco, L.; Sorbi, S.; Nacmias, B. KIBRA T allele influences memory performance and progression of cognitive decline: A 7-year follow-up study in subjective cognitive decline and mild cognitive impairment. Neurol. Sci. 2019, 40, 1559–1566. [Google Scholar] [CrossRef] [PubMed]
- Pamio, M.V.; Trevisan, C.; Pigozzo, S.; De Rui, M.; Devita, M.; Girardi, A.; Manzato, E.; Sergi, G.; Coin, A. Are cytochrome P4502D6 and apolipoprotein E genotypes associated with long-term cognitive and functional changes in patients treated with donepezil? Psychogeriatrics 2020, 20, 578–584. [Google Scholar] [CrossRef] [PubMed]
- Piccardi, M.; Congiu, D.; Squassina, A.; Manconi, F.; Putzu, P.F.; Mereu, R.M.; Chillotti, C.; Del Zompo, M. Alzheimer’s disease: Case-control association study of polymorphisms in ACHE, CHAT, and BCHE genes in a Sardinian sample. Am. J. Med Genet. Part B Neuropsychiatr. Genet. 2007, 144, 895–899. [Google Scholar] [CrossRef] [PubMed]
- Poli, M.; Gatta, L.B.; Dominici, R.; Lovati, C.; Mariani, C.; Albertini, A.; Finazzi, D. Apolipoprotein E haplotyping by denaturing high-performance liquid chromatography. Clin. Chem. Lab. Med. 2005, 43, 512–518. [Google Scholar] [CrossRef]
- Polito, L.; Kehoe, P.G.; Davin, A.; Benussi, L.; Ghidoni, R.; Binetti, G.; Quadri, P.; Lucca, U.; Tettamanti, M.; Clerici, F.; et al. The SIRT2 polymorphism rs10410544 and risk of Alzheimer’s disease in two Caucasian case–control cohorts. Alzheimer’s Dement. 2013, 9, 392–399. [Google Scholar] [CrossRef] [PubMed]
- Pozzi, F.E.; Conti, E.; Appollonio, I.; Ferrarese, C.; Tremolizzo, L. Predictors of response to acetylcholinesterase inhibitors in dementia: A systematic review. Front. Neurosci. 2022, 16, 998224. [Google Scholar] [CrossRef]
- Rademakers, R.; Baker, M.; Gass, J.; Adamson, J.; Huey, E.D.; Momeni, P.; Spina, S.; Coppola, G.; Karydas, A.M.; Stewart, H.; et al. Phenotypic variability associated with progranulin haploinsufficiency in patients with the common 1477C→T (Arg493X) mutation: An international initiative. Lancet Neurol. 2007, 6, 857–868. [Google Scholar] [CrossRef]
- de Matos, M.R.; Ferreira, C.; Herukka, S.-K.; Soininen, H.; Janeiro, A.; Santana, I.; Baldeiras, I.; Almeida, M.R.; Lleó, A.; Dols-Icardo, O.; et al. Quantitative Genetics Validates Previous Genetic Variants and Identifies Novel Genetic Players Influencing Alzheimer’s Disease Cerebrospinal Fluid Biomarkers. J. Alzheimer’s Dis. 2018, 66, 639–652. [Google Scholar] [CrossRef]
- Serretti, A.; Olgiati, P.; De Ronchi, D. Genetics of Alzheimer’s Disease. A Rapidly Evolving Field. J. Alzheimer’s Dis. 2007, 12, 73–92. [Google Scholar] [CrossRef] [PubMed]
- Weiner, M.W.; Veitch, D.P.; Aisen, P.S.; Beckett, L.A.; Cairns, N.J.; Cedarbaum, J.; Green, R.C.; Harvey, D.; Jack, C.R.; Jagust, W.; et al. 2014 Update of the Alzheimer’s Disease Neuroimaging Initiative: A review of papers published since its inception. Alzheimer’s Dement. 2015, 11, e1–e120. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Yao, Z.; Jiang, Y.; Keller, E.T. Prostate cancer stem cell biology. Minerva Urol. E Nefrol. Ital. J. Urol. Nephrol. 2012, 64, 19–33. [Google Scholar]
- van der Maaten LJ, P.; Hinton, G.E. Visualizing high-dimensional data using t-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]
- Haug, C.J.; Drazen, J.M. Artificial Intelligence and Machine Learning in Clinical Medicine, 2023. N. Engl. J. Med. 2023, 388, 1201–1208. [Google Scholar] [CrossRef]
- Abrego-Guandique, D.M.; Bonet, M.L.; Caroleo, M.C.; Cannataro, R.; Tucci, P.; Ribot, J.; Cione, E. The Effect of Beta-Carotene on Cognitive Function: A Systematic Review. Brain Sci. 2023, 13, 1468. [Google Scholar] [CrossRef]
- Bevacqua, E.; Ammirato, S.; Cione, E.; Curcio, R.; Dolce, V.; Tucci, P. The Potential of MicroRNAs as Non-Invasive Prostate Cancer Biomarkers: A Systematic Literature Review Based on a Machine Learning Approach. Cancers 2022, 14, 5418. [Google Scholar] [CrossRef]
- Kim, D.; Im, T. A Systematic Review of Virtual Reality-Based Education Research Using Latent Dirichlet Allocation: Focus on Topic Modeling Technique. Mob. Inf. Syst. 2022, 2022, 1201852. [Google Scholar] [CrossRef]
- Mo, Y.; Kontonatsios, G.; Ananiadou, S. Supporting systematic reviews using lda-based document representations. Syst. Rev. 2015, 4, 172. [Google Scholar] [CrossRef]
- Bilro, R.; Loureiro, S.; Souto, A. A systematic review of customer behavior in business-to-business markets and agenda for future research. J. Bus. Ind. Mark. 2023, 38, 122–142. [Google Scholar] [CrossRef]
- Lenci, A. Distributional semantics in linguistic and cognitive research. Ital. J. Linguist. 2008, 20, 1–31. [Google Scholar]
- Gallelli, L.; Cione, E.; Peltrone, F.; Siviglia, S.; Verano, A.; Chirchiglia, D.; Zampogna, S.; Guidetti, V.; Sammartino, L.; Montana, A.; et al. Hsa-miR-34a-5p and hsa-miR-375 as Biomarkers for Monitoring the Effects of Drug Treatment for Migraine Pain in Children and Adolescents: A Pilot Study. J. Clin. Med. 2019, 8, 928. [Google Scholar] [CrossRef] [PubMed]
- Perri, M.; Lucente, M.; Cannataro, R.; De Luca, I.F.; Gallelli, L.; Moro, G.; De Sarro, G.; Caroleo, M.C.; Cione, E.; Cafiero, C. Variation in Immune-Related microRNAs Profile in Human Milk Amongst Lactating Women. MicroRNA 2018, 7, 107–114. [Google Scholar] [CrossRef] [PubMed]










| Name, Year | Ref | Gene | Mutations | Localization | Other |
|---|---|---|---|---|---|
| Albani et al., 2012 | [27] | --------------------------- | ----------------------------------------- | ---------------- | Not in Alzheimer |
| Andreoli et al., 2014 | [28] | GRINB2 | rs7301328 (GRINB2), rs1805482 (GRINB2), rs3026160 (GRINB2), rs1806201 (GRINB2), rs1806191 (GRINB2) | Italy | Alzheimer |
| Bagnoli et al., 2007 | [29] | IL-10 | rs1800896 (IL-10), rs1800871 (IL-10) | Italy | Alzheimer |
| Bagnoli et al., 2013 | [30] | TOMM40 | rs157580 (TOMM40), rs2075650 (TOMM40), rs15758 (TOMM40) | Italy | Alzheimer |
| Bartoletti-Stella et al., 2022 | [31] | ELAVL1, EP300, EPHA1, FERMT2, INPP5D, MARK2, MARK4, PICALM PLCG2, PTK2B, RIN3, TOMM40 ZCWPW1, ADAM10, BIN1, CLU, CR1 | c.112A > G (ADAM10), c.556dupC (ADAM10), c.696C > A (BIN1), c.865G > A (BIN1), c.1462-3C > T (BIN1), c.509C > T (CLU), c.4956G > A (CR1), c.4356T > C (CR1), c.765C > T (ELAVL1), c.2194C > T (EP300), c.928A > G (EPHA1), c.1077G > C (FERMT2), c.1538C > T (FERMT2), c.470G > A (INPP5D), c.2085C > T (INPP5D), c.1611C > T (MARK2), c.1553C > T (MARK4), c.1231G > C (PICALM), c.3379C > A (PLGC2), c.408G > A (PLCG2), c.2591C > T (PTK2B), c.2377T > C (RIN3),c.384C > G (TOMM40), c.1834C > T (ZCWPW1), c.314A > G (ZCWPW1), c.283-5T > G (ZCWPW1) | Italy | Alzheimer |
| Belloy et al., 2022 | [32] | ApoE | rs439401 (ApoE) | Not in Italy | Alzheimer |
| Bizzarro et al., 2009 | [33] | ApoE | rs449647 (ApoE), rs405509 (ApoE), rs769446 (ApoE), rs429358 (ApoE) rs7412 (ApoE) | Italy | Alzheimer |
| Bosco et al., 2013 | [34] | ----------------------------- | ------------------------------------------ | ---------- | Review |
| Broer et al., 2015 | [35] | ----------------------------- | ------------------------------------------ | ---------------- | Not Alzheimer |
| Bucossi et al., 2012 | [36] | ATP7B | rs1061472 (ATP7B), rs732774 (ATP7B) | Italy | Alzheimer |
| Capurso et al., 2010 | [37] | IL-6 | –174 G/C (IL-6) | Italy | Alzheimer |
| Capurso et al., 2010 | [38] | GSTO1, APOE | rs7412 (ApoE), rs429358 (ApoE), rs4925 (GSTO1), rs1804834 (GSTO1) | Italy | Alzheimer |
| Cellini et al., 2009 | [39] | SORL1 | rs661057 (SORL1), rs11218304 (SORL1), rs560573 (SORL1), rs12364988 (SORL1), rs668387 (SORL1), rs689021 (SORL1), rs641120 (SORL1), rs556349 (SORL1), rs2070045 (SORL1), rs1699102 (SORL1), rs3824968 (SORL1), rs2282649 (SORL1), rs1010159 (SORL1) | Italy | Alzheimer |
| Ciminelli et al., 2020 | [40] | AKR7A2, ALDH5A1, ABAT | rs4646832 (ALDH5A1), rs4646828 (ALDH5A1), rs2760118 (ALDH5A1), rs3765310 (ALDH5A1), rs1043657 (AKR7A2), rs1731017 (ABAT) | Italy | Alzheimer |
| Clarelli et al., 2016 | [41] | CHRNA7 | rs6494223 (CHRNA7), rs8024987 (CHRNA7) | Italy | Alzheimer |
| Colacicco et al.,2009 | [42] | A2M, ORL1 | rs669 (A2M), +1073 (ORL1) +1071 (ORL1) | Italy | Alzheimer |
| Corbo et al.,2009 | [43] | CYP19 | rs12907866 (CYP19), rs17601241 (CYP19), rs4646 (CYP19) | Italy | Alzheimer |
| Cruz-Sanabria et al., 2021 | [44] | ----------------------------- | ------------------------------------------ | -------------- | Not Alzheimer |
| DeMichele-Sweet et al., 2021 | [45] | ENPP6, SUMF1 | rs9994623 (ENPP6), rs201109606 (SUMF1) | Not in Italy | Alzheimer |
| Di Maria et al., 2012 | [46] | NGF | rs6330 (NGF), rs11466110 (NGF), rs11466111 (NGF), rs6325 (NGF), rs35941329 (NGF) | Italy | Alzheimer |
| Emanuele et al., 2011 | [47] | CDKN2B-AS1 | rs1333049 (CDKN2B-AS1) | Italy | Alzheimer |
| Finckh et al., 2003 | [48] | PLAU | −141 C/T (PLAU) | Italy | Alzheimer |
| Fortney et al., 2015 | [49] | ----------------------------- | ----------------------------------------- | ------------ | Not alzheimer |
| Galimberti et al., 2008 | [50] | NOS 1 | Ex1f-VNTR (NOS1) | Italy | Alzheimer |
| Guerini et al., 2016 | [51] | SNAP-25 | rs363050 (SNAP-25), rs363039 (SNAP-25), rs363043 (SNAP-25) | Italy | Alzheimer |
| Hollingworth et al., 2011 | [52] | CUX2, CDK1, CNTN5, C16orf88, IQCK, MS4A6A, CR1, MS4A6A, BIN1, ABCA7 | rs3764650 (ABCA7), rs744373 (BIN1), rs670139 (MS4A4E), rs3818361 (CR1), rs610932 (MS4A6A), rs7191155 (IQCK), rs4782279 (IQCK), rs1858973 (IQCK), rs739565 (C16orf88), rs10501927 (CNTN5), Rs10761558 (CDK1), rs3809278 (CUX2) | Italy | Alzheimer |
| Jun et al., 2016 | [53] | ARL17B | rs2732703 (ARL17B) | NR | Alzheimer |
| Lambert et al., 2009 | [54] | CLU, CR1 | rs11136000 (CLU), rs2279590 (CLU), rs9331888 (CLU), rs6656401 (CR1), rs3818361 (CR1) | Italy | Alzheimer |
| Lambert et al., 2013 | [55] | BIN1, HLA-DRB5/HLA-DRB1, ZCWPW1, PTK2B, CELF1, MS4A6A | rs6733839 (BIN1), rs9271192 (HLA-DRB5/HLA-DRB1), rs1476679 (ZCWPW1), rs28834970 (PTK2B), rs9331896 (CLU), rs10838725 (CELF1), rs983392 (MS4A6A) | Italy | Alzheimer |
| Lanni et al., 2012 | [56] | COMT | rs4680 (COMT) | Italy | Alzheimer |
| Laws et al., 2005 | [57] | TNF | rs1799724 (TNF), rs1800629 (TNF) | Not in Italy | Alzheimer |
| Lescai et al., 2010 | [58] | PCDH11X | rs5984894 (PCDH11X) | Italy | Alzheimer |
| Lescai et al., 2011 | [59] | ApoE | rs449647 (ApoE), rs769446 (ApoE), rs405509 (ApoE), rs429358 (ApoE), rs7412 (ApoE) | Italy | Alzheimer |
| Licastro et al., 2011 | [60] | IL10, TNF, IL6, IFNG [SERPINA3, HMGCR | –1082G/A(IL10), –308 G/A(TNF), −174 G/C (IL6), +874 T/A (IFNG), –51 G/T (SERPINA 3), –911 C/A(HMGCR) | Italy | Alzheimer |
| Licastro et al., 2015 | [61] | IL-28 b, Med23 | rs12979860 (IL-28 B), rs3756784 (MED 23) | Italy | Alzheimer |
| Lio et al., 2003 | [62] | IL-10 | −1082 G/A (IL-10), −819 C/T (IL-10), −592 C/A (IL-10) | Italy | Alzheimer |
| Lio et al., 2006 | [63] | TNF-α | −308 G/A (TNF-α) | Italy | Alzheimer |
| Lu et al., 2017 | [64] | HLA-DRB1 | rs9271192 (HLA-DRB1) | Not in Italy | Alzheimer |
| Lupton et al., 2016 | [65] | TREM2 | rs9394721 (TREM2) | Not in Italy | Alzheimer |
| Maletta et al., 2018 | [66] | MTHFR, ACE, AGT, CYP7A1, PAI-1, FII G20210A, FV HR2 H1299R, FV Leiden, GPIIIa, CETP | rs1801133 (MTHFR), rs1801131 (MTHFR), rs1799752 (ACE), rs699 (AGT), 844ins68 (CBS),rs3808607 (CYP7A1), rs1799889 (PAI-1), rs1799963 (FII G20210A), rs1800595 (FV HR2 H1299R), rs6025 (FV Leiden), rs5918 (GPIIIa), rs5882 (CETP) | Italy | Alzheimer |
| Mariani et al., 2013 | [67] | HFE, TF, C282Y | rs1800562 (C282Y), rs1799945 (HFE), rs1049296 (TF) | Italy | Alzheimer |
| Masri et al., 2020 | [68] | PICALM | rs3851179G > A (PICALM) | Not in Italy | Alzheimer |
| Minoretti et al., 2006 | [69] | TLR4 | Asp299Gly (TLR4) | Italy | Alzheimer |
| Montesanto et al., 2016 | [70] | UCP2, UCP3, UCP4, UCP5 | rs2306820 (UCP3), rs655717 (UCP2), rs660339 (UCP2), rs659366 (UCP2), rs635441 (UCP2), rs1685354 (UCP3), rs2734827 (UCP3), rs1800849 (UCP3), rs10498769 (UCP4), rs12192544 (UCP4), rs3757241 (UCP4), rs9472817 (UCP4), rs17314910 (UCP5), rs6637742 (UCP5), rs3007756 (UCP5), rs5930414 (UCP5) | Italy | Alzheimer |
| Nacmias et al., 2009 | [71] | GAB2 | rs2373115 (GAB2) | Italy | Alzheimer |
| Napolioni et al., 2011 | [72] | ----------------------------- | ----------------------------------------- | ---------------- | Not Alzheimer |
| Olgiati et al., 2013 | [73] | SORL1 | rs668387 (SORL1), rs68902 (SORL1), rs641120 (SORL1) | NR | Alzheimer |
| Orlacchio et al., 2002 | [74] | NCSTN | 237 G/A (NCSTN) 747 C/T (NCSTN) | Italy | Alzheimer |
| Pilotto et al., 2009 | [75] | CYP2D6 | rs1080985 (CYP2D6) | Italy | Alzheimer |
| Pola et al., 2004 | [76] | MCP-1 | −2518 A/G (MCP-1) | Italy | Alzheimer |
| Poleggi et al., 2008 | [77] | PRNP | Met129- Val (PRNP) | Italy | Alzheimer |
| Poli et al., 2008 | [78] | APH-1b | C + 651T > G (APH-1b) | Italy | Alzheimer |
| Porrello et al., 2006 | [79] | Er-α | PvuII (−397 T/C), XbaI (−351 A/G) (Er-α) | Italy | Alzheimer |
| Scacchi et al., 2009 | [80] | P53, P73 | rs1042522 (P53), rs2273953 (P73), rs1801173 (P73), rs3765728 (P73), rs1801174 (P73) | Italy | Alzheimer |
| Scasellati et al., 2004 | [81] | IL-10 | −1082 G/A (IL-10), −819 T/C (IL-10), −592 C/A (IL-10) | Italy | Alzheimer |
| Schmidt et al., 2011 | [82] | ----------------------------- | ----------------------------------------- | ---------------- | Not in Alzheimer |
| Schott et al., 2016 | [83] | CR1, BIN1, INPP5D, SCARB2, SNCA, MEF2C, CD2AP, EPHA1, NME8, ZCWPW, CLU, PTK2B, CELF1, MS4A4E, PICALM, SORL1, FERMT2, SLC4A4, RIN3, ABCA7, APOE, CD33, EXOC3L2, BLOC1S3, MARK4, CASS4 | rs3818361 (CR1), rs744373 (BIN1), rs35349669 (INPP5D), rs6825004 (SCARB2), rs7687945 (SNCA), rs190982 (MEF2C), rs10948363 (CD2AP), rs11767557 (EPHA1), rs2718058 (NME8), rs1476679 (ZCWPW), rs11136000 (CLU), rs28834970 (PTK2B), rs10838725 (CELF1), rs10838725 (MS4A4E), rs10838725 (PICALM), rs670139 (SORL1), rs983392 (FERMT2), rs3851179 (SLC4A4), rs11218343 (RIN3), rs17125944 (ABCA7), rs10498633 (APOE), rs3764650 (CD33), rs2075650 (EXOCL3L2), rs3865444 (BLOC1S3),rs597668 (MARK4), rs7274581 (CAS4) | Not only in Italy | Alzheimer |
| Scola et al., 2003 | [84] | IFN-γ | +874T→A (IFN-γ) | Italy | Alzheimer |
| Seripa et al., 2008 | [85] | GRIN2B | rs1019385 (GRIN2B), rs1806201 (GRIN2B), rs890 (GRIN2B) | Italy | Alzheimer |
| Seripa et al., 2008 | [86] | RELN, LIMK2 | rs607755 (RELN), rs2229864 (LIMK2) | Italy | Alzheimer |
| Seripa et al., 2011 | [87] | ----------------------------- | ------------------------------------------ | ---------------- | Review |
| Serpente et al., 2011 | [88] | ORL1 | rs1050283 (ORL1) | Italy | Alzheimer |
| Squillario et al., 2020 | [89] | TOMM40, GRM7 | rs2075650 (TOMM40), rs8106922 (TOMM40), rs9311976 (GRM7), rs266410 (GRM7) | NR | Alzheimer |
| Talwar et al., 2021 | [90] | ApoE, EGFR, ACTB | rs405509 (ApoE), rs7259620 (ApoE), rs769449 (ApoE), rs725617 (ApoE), rs7256173 (ApoE), rs6970262 (EGFR), rs852423 (ACTB) | Not in Italy | Alzheimer |
| Tedde et al., 2010 | [91] | NEDD9 | rs760678 (NEDD9) | Italy | Alzheimer |
| Tindale et al., 2017 | [92] | ----------------------------- | ------------------------------------------ | ---------------- | Not Alzheimer |
| Tisato et al., 2018 | [93] | TF, HFE, FPN1, HAMP | HFE C282Y, HFE H63D, FPN1 −8CG, HAMP −582AG, TF P570S | Italy | Alzheimer |
| Valenza et al., 2010 | [94] | ApoE | −491 A/T (ApoE) | Italy | Alzheimer |
| Venturelli et al., 2005 | [95] | eNOS | T-786C (eNOS) | Italy | Alzheimer |
| Wang et al., 2016 | [96] | GAB2, PICALM, SORL1 | rs1010159 (SORL1), rs12285364 (SORL1), rs1699102 (SORL1), rs2070045 (SORL1), rs2282649 (SORL1), rs3824968 (SORL1), rs4935774 (SORL1), rs556349 (SORL1), rs641120 (SORL1), rs661057 (SORL1), rs668387 (SORL1), rs689021 (SORL1), rs3851179 (PICALM), rs541458 (PICALM), rs2373115 (GAB2) | Not in Italy | Alzheimer |
| Name, Year | Ref | Gene | Mutations | Localization | Other |
|---|---|---|---|---|---|
| Bernardi et al., 2012 | [97] | ------------------------------ | -------------------------------------- | --------------------- | Not Alzheimer |
| Calabretta et al., 2009 | [98] | ------------------------------ | -------------------------------------- | --------------------- | Not Alzheimer |
| Cheng et al., 2012 | [99] | NGFR | rs734194 (NGFR), rs2072445 (NGFR), rs2072446 (NGFR), rs741072 (NGFR), rs741073 (NGFR) | Not in Italy | Alzheimer |
| De Rojas et al., 2021 | [100] | PRKD3/NDUFAF7, SHARPIN, PLCG2, CHRNE, APP | rs876461 (PRKD3/NDUFAF7), rs34674752 (SHARPIN), rs34173062 (SHARPIN), rs3935877 (PLCG2), rs72835061 (CHRNE), rs2154481 (APP) | NR | Alzheimer |
| Flex et al., 2006 | [101] | MMP-1, MMP-3, MMP-9 | 2G2G (MMP-1), 1G2G (MMP-1), 5A5A (MMP-3), TT (MMP-9) | Italy | Alzheimer |
| Grunbatt et al., 2011 | [102] | CHAT | rs10776585 (CHAT), rs2289305 (CHAT), rs3750752 (CHAT), rs12356649 (CHAT), rs11591558 (CHAT), rs8178984 (CHAT), rs1153783 (CHAT), rs1880676 (CHAT), rs12359885 (CHAT), rs8178990 (CHAT), rs10082479 (CHAT), rs17775758 (CHAT), rs7903612 (CHAT), rs7091005 (CHAT), rs2889759 (CHAT), rs4838391 (CHAT), rs11101186 (CHAT), rs10857520 (CHAT) | Italy | Alzheimer |
| Hansmannel et al., 2010 | [103] | OTC | rs5963409 (OTC), rs5963411 (OTC) | Italy | Alzheimer |
| Hong et al., 2020 | [104] | ApoE, LINC01570, TOMM40, LOC124903153, NECTIN 2, LOC107985236, LINC01030, LINC00624, APOC1, ADAMTS3, ADGRB3, FZD4-DT, LOC107984589, SLC2A3, NMNAT2 | rs429358 (ApoE), rs10400961 (LINC01570), rs34095326 (TOMM40), rs1004954 (LOC124903153), rs34794167 (LOC124903153), rs6857 (NECTIN2), rs12857276 (LOC124903153), rs7254133 (GRCh38?), rs2209929 (LOC124903153), rs3814341 (LOC107985236), rs2023625 (LINC01030), rs35386129 (LOC124903153), rs2321744 (LOC124903153), rs985017 (LOC124903153), rs769449 (ApoE), rs985018 (LOC124903153), rs36015381 (LOC124903153), rs34528363 (LOC124903153), rs12972970 (NECTIN2), rs12972156 (NECTIN2), rs68058618 (GRCh38?), rs12429500 (LOC124903153), rs6672481 (LINC00624), rs2353964 (LINC00624), rs10900366 (LINC00624), rs12721051 (APOC1), rs12408395 (LINC00624), rs9592217 (LOC124903153), rs9598510 (LOC124903153), rs10900364 (LINC00624), rs11240038 (LINC00624),rs9598511 (LOC124903153), rs6657402 (LINC00624), rs2353972 (LINC00624), rs36055662 (ADAMTS3), rs117950083 (ADGRB3) rs35709873 (LOC124903153), rs56131196 (APOC1), rs4420638 (APOC1), rs55696402 (LOC124903153), rs11605035 (FZD4-DT), rs11606999 (FZD4-DT), rs11607037 (FZD4-DT),rs10898566 (FZD4-DT), rs10898565 (FZD4-DT),rs12999830 (GRCh38), rs11240037 (GRCh38), rs11240043 (LINC00624), rs2075650 (TOMM40), rs34404554 (TOMM40), rs4600098 (LINC00624), rs6684440 (LINC00624), rs4381253 (LINC00624), rs6683579 (LINC00624), rs6657222 (LINC00624), rs2883315 (LINC00624), rs4339909 (LINC00624), rs6696385 (LINC00624), rs11581799 (LINC00624), rs11556505 (TOMM40), rs4375326 (LINC00624), rs11576468 (LINC00624), rs11589438 (LINC00624), rs12742771 (LINC00624), rs11240050 (LINC00624), rs78581038 (LOC107984589), rs12812878 (SLC2A3), rs6695692 (LINC00624), rs6688033 (LINC00624), rs11234901 (FZD4-DT), rs6688244 (LINC00624), rs6678706 (LINC00624), rs11605694 (FZD4-DT),rs58619449 (FZD4-DT), rs58370115 (FZD4-DT), rs7127641 (FZD4-DT), rs61809265 (NMNAT2), rs35519936 (LOC124903153), rs34342646 (NECTIN2),rs10414043 (APOC1), rs72639166 (FZD4-DT),rs7256200 (APOC1) | Not in Italy | Alzheimer |
| Krumbiegel et al., 2010 | [105] | ------------------------------ | -------------------------------------- | --------------------- | Not Alzheimer |
| Lambert et al., 2011 | [106] | BIN1, EXO3CL2, PICALM | rs744373 (BIN1), rs597668 (EXO3CL2), rs541458 (PICALM) | Italy | Alzheimer |
| Lambert et al., 2013 | [107] | FRMD4A | rs7081208 (FRMD4A), rs2446581 (FRMD4A), rs17314229 (FRMD4A) | Italy | Alzheimer |
| Mazzeo et al., 2019 | [108] | ------------------------------ | -------------------------------------- | --------------------- | Not Alzheimer |
| Pamio et al., 2020 | [109] | CYP2D6 | rs1080985 (CYP2D6) | Italy | Alzheimer |
| Piccardi et al., 2007 | [110] | CHAT, AChE | rs2177369 (CHAT), rs12705094 (AChE), rs3087504 (AChE), rs3757869 (AChE) | Italy | Alzheimer |
| Poli et al., 2005 | [111] | ApoE | rs11542041 (ApoE), rs11542035 (ApoE), rs769455 (ApoE) | Italy | Not Alzheimer |
| Polito et al., 2013 | [112] | SIRT2, SIRT3 | rs10410544 (SIRT2), rs4980329(SIRT3), rs536715 (SIRT3), rs2015 (SIRT2), rs3825075 (SIRT3), rs11880757 (SIRT2), rs11879010 (SIRT2), rs11667030 (SIRT2) | Italy | Alzheimer |
| Pozzi et al., 2022 | [113] | ------------------------------ | -------------------------------------- | --------------------- | SR |
| Rademakers et al., 2007 | [114] | GRN, MAPT | Arg493X (GRN), rs1052553 (MAPT), rs7412 (ApoE), rs429358 (ApoE), rs4792937(GRN), rs2879096 (GRN), c.–7–320G→C (GRN), rs34424835 (GRN), rs9897528 (GRN), rs25646 (GRN), c.835 + 7A→G (GRN), rs5848 (GRN), rs34424835 (GRN) | Not in Italy | Alzheimer |
| Ramos De Matos et al., 2018 | [115] | APOE, LOC100129500, PVRL2, SNAR-I, TOMM40, INPP5, CD2AP, GLIS3, PVRL2, CASS4 | rs35349669(INPP5), rs1316356 (SNAR-1), rs9877502 (SNAR-1), rs9349407 (CD2AP), rs514716 (GLIS3), rs12972156 (PVRL2), rs34342646 (PVRL2), rs71352238 (TOMM40),rs157580 (TOMM40), rs2075650 (TOMM40), rs34404554 (TOMM40), rs11556505 (TOMM40), rs769449 (APOE), rs429358 (APOE), rs439401 (LOC100129500), rs7274581 (CASS4) | Italy * | Alzheimer |
| Serretti et al., 2007 | [116] | ------------------------------ | -------------------------------------- | --------------------- | Review |
| Weiner et al., 2015 | [117] | ------------------------------ | -------------------------------------- | --------------------- | Review |
| Yu et al., 2012 | [118] | ------------------------------ | -------------------------------------- | --------------------- | Not Alzheimer |
| Name, Year | Ref. | Gene | Mutations | Localization |
|---|---|---|---|---|
| Bizzarro et al., 2009 | [33] | ApoE | rs449647, rs405509, rs769446 | Italy |
| Capurso et al., 2010 | [38] | ApoE | rs7412, rs429358 | Italy |
| Lescai et al., 2011 | [59] | ApoE | rs449647, rs769446, rs405509, rs429358, rs7412 | Italy |
| Valenza et al., 2010 | [94] | ApoE | −491 AT vs −491 AA | Italy |
| Ramos de matos et al., 2018 | [115] | ApoE | rs769449, rs429358 | Italy * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saraceno, G.F.; Abrego-Guandique, D.M.; Cannataro, R.; Caroleo, M.C.; Cione, E. Machine Learning Approach to Identify Case-Control Studies on ApoE Gene Mutations Linked to Alzheimer’s Disease in Italy. BioMedInformatics 2024, 4, 600-622. https://doi.org/10.3390/biomedinformatics4010033
Saraceno GF, Abrego-Guandique DM, Cannataro R, Caroleo MC, Cione E. Machine Learning Approach to Identify Case-Control Studies on ApoE Gene Mutations Linked to Alzheimer’s Disease in Italy. BioMedInformatics. 2024; 4(1):600-622. https://doi.org/10.3390/biomedinformatics4010033
Chicago/Turabian StyleSaraceno, Giorgia Francesca, Diana Marisol Abrego-Guandique, Roberto Cannataro, Maria Cristina Caroleo, and Erika Cione. 2024. "Machine Learning Approach to Identify Case-Control Studies on ApoE Gene Mutations Linked to Alzheimer’s Disease in Italy" BioMedInformatics 4, no. 1: 600-622. https://doi.org/10.3390/biomedinformatics4010033