Modified ECM-Based Bioink for 3D Printing of Multi-Scale Vascular Networks
Abstract
:1. Introduction
2. Results and Discussion
2.1. Fabrication of the YIGSR-ECM Hydrogel
2.2. Characterization of the YIGSR-ECM Hydrogel
2.3. The 2D and 3D Culture of iPSC-ECs
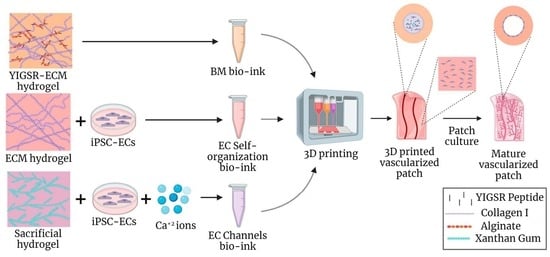
2.4. The 3D Bioprinting of Vascularized Patches
3. Conclusions
4. Materials and Methods
4.1. Materials
4.2. Methods
4.2.1. ECM Hydrogel Production
4.2.2. YIGSR-LF200 Alginate Production
4.2.3. YIGSR-ECM Hydrogel Production
4.2.4. YIGSR-ECM Hydrogel Crosslinking
4.2.5. Rheological Properties
4.2.6. X-ray Photoelectron Spectroscopy (XPS)
4.2.7. Spectrophotometry
4.2.8. HUVECs Culture
4.2.9. Induced Pluripotent Stem Cell (iPSC) Culture
4.2.10. EC Differentiation from iPSCs
4.2.11. Flow Cytometry
4.2.12. The 3D Cell Encapsulation in Hydrogels
4.2.13. Proliferation Assay
4.2.14. Viability Assay
4.2.15. The 2D Cell Seeding on Hydrogels
4.2.16. Immunofluorescence Staining
4.2.17. High-Resolution Scanning Electron Microscopy (hrSEM)
4.2.18. Hydrolytic and Enzymatic Swelling/Degradation Assay
4.2.19. Bio-Inks Preparation
4.2.20. Vascular Patch Printing Process
4.2.21. Statistical Analysis
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Novosel, E.C.; Kleinhans, C.; Kluger, P.J. Vascularization Is the Key Challenge in Tissue Engineering. Adv. Drug Deliv. Rev. 2011, 63, 300–311. [Google Scholar] [CrossRef] [PubMed]
- Song, H.H.G.; Rumma, R.T.; Ozaki, C.K.; Edelman, E.R.; Chen, C.S. Vascular Tissue Engineering: Progress, Challenges, and Clinical Promise. Cell Stem Cell 2018, 22, 340–354. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, C.; Brady, E.; Zheng, Y.; Moore, E.; Stevens, K.R. Engineering the Multiscale Complexity of Vascular Networks. Nat. Rev. Mater. 2022, 7, 702–716. [Google Scholar] [CrossRef] [PubMed]
- Rademakers, T.; Horvath, J.M.; van Blitterswijk, C.A.; LaPointe, V.L.S. Oxygen and Nutrient Delivery in Tissue Engineering: Approaches to Graft Vascularization. J. Tissue Eng. Regen. Med. 2019, 13, 1815–1829. [Google Scholar] [CrossRef] [PubMed]
- Pugsley, M.K.; Tabrizchi, R. The Vascular System: An Overview of Structure and Function. J. Pharmacol. Toxicol. Methods 2000, 44, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Cleaver, O.; Melton, D.A. Endothelial Signaling during Development. Nat. Med. 2003, 9, 661–668. [Google Scholar] [CrossRef] [PubMed]
- Camasão, D.B.; Mantovani, D. The Mechanical Characterization of Blood Vessels and Their Substitutes in the Continuous Quest for Physiological-Relevant Performances. A Critical Review. Mater. Today Bio 2021, 10, 100106. [Google Scholar] [CrossRef]
- Sánchez-Molina, D.; García-Vilana, S.; Llumà, J.; Galtés, I.; Velázquez-Ameijide, J.; Rebollo-Soria, M.C.; Arregui-Dalmases, C. Mechanical Behavior of Blood Vessels: Elastic and Viscoelastic Contributions. Biology 2021, 10, 831. [Google Scholar] [CrossRef]
- Wettschureck, N.; Strilic, B.; Offermanns, S. Passing the Vascular Barrier: Endothelial Signaling Processes Controlling Extravasation. Physiol. Rev. 2019, 99, 1467–1525. [Google Scholar] [CrossRef]
- Hallmann, R.; Horn, N.; Selg, M.; Wendler, O.; Pausch, F.; Sorokin, L.M. Expression and Function of Laminins in the Embryonic and Mature Vasculature. Physiol. Rev. 2005, 85, 979–1000. [Google Scholar] [CrossRef]
- Candiello, J.; Balasubramani, M.; Schreiber, E.M.; Cole, G.J.; Mayer, U.; Halfter, W.; Lin, H. Biomechanical Properties of Native Basement Membranes. FEBS J. 2007, 274, 2897–2908. [Google Scholar] [CrossRef] [PubMed]
- Kubota, Y.; Kleinman, H.K.; Martin, G.R.; Lawley, T.J. Role of Laminin and Basement Membrane in the Morphological Differentiation of Human Endothelial Cells into Capillary-like Structures. J. Cell Biol. 1988, 107, 1589–1598. [Google Scholar] [CrossRef] [PubMed]
- Leclech, C.; Natale, C.F.; Barakat, A.I. The basement membrane as a structured surface–role in vascular health and disease. J. Cell Sci. 2020, 133, jcs239889. [Google Scholar] [CrossRef] [PubMed]
- Yadid, M.; Oved, H.; Silberman, E.; Dvir, T. Bioengineering Approaches to Treat the Failing Heart: From Cell Biology to 3D Printing. Nat. Rev. Cardiol. 2021, 19, 83–99. [Google Scholar] [CrossRef] [PubMed]
- Miyagi, Y.; Chiu, L.L.Y.; Cimini, M.; Weisel, R.D.; Radisic, M.; Li, R.K. Biodegradable Collagen Patch with Covalently Immobilized VEGF for Myocardial Repair. Biomaterials 2011, 32, 1280–1290. [Google Scholar] [CrossRef] [PubMed]
- Anderson, E.M.; Silva, E.A.; Hao, Y.; Martinick, K.D.; Vermillion, S.A.; Stafford, A.G.; Doherty, E.G.; Wang, L.; Doherty, E.J.; Grossman, P.M.; et al. VEGF and IGF Delivered from Alginate Hydrogels Promote Stable Perfusion Recovery in Ischemic Hind Limbs of Aged Mice and Young Rabbits. J. Vasc. Res. 2017, 54, 288–298. [Google Scholar] [CrossRef] [PubMed]
- Lesman, A.; Habib, M.; Caspi, O.; Gepstein, A.; Arbel, G.; Levenberg, S.; Gepstein, L. Transplantation of a Tissue-Engineered Human Vascularized Cardiac Muscle. Tissue Eng. Part A 2009, 16, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Redenski, I.; Landau, S.; Szklanny, A.; Merdler, U.; Levenberg, S. Prevascularized Scaffolds Bearing Human Dental Pulp Stem Cells for Treating Complete Spinal Cord Injury. Adv. Health Mater. 2020, 9, 2000974. [Google Scholar] [CrossRef]
- Andrée, B.; Ichanti, H.; Kalies, S.; Heisterkamp, A.; Strauß, S.; Vogt, P.M.; Haverich, A.; Hilfiker, A. Formation of Three-Dimensional Tubular Endothelial Cell Networks under Defined Serum-Free Cell Culture Conditions in Human Collagen Hydrogels. Sci. Rep. 2019, 9, 5437. [Google Scholar] [CrossRef]
- Klotz, B.J.; Oosterhoff, L.A.; Utomo, L.; Lim, K.S.; Vallmajo-Martin, Q.; Clevers, H.; Woodfield, T.B.F.; Rosenberg, A.J.W.P.; Malda, J.; Ehrbar, M.; et al. A Versatile Biosynthetic Hydrogel Platform for Engineering of Tissue Analogues. Adv. Health Mater. 2019, 8, 1900979. [Google Scholar] [CrossRef]
- Landau, S.; Guo, S.; Levenberg, S. Localization of Engineered Vasculature within 3D Tissue Constructs. Front. Bioeng. Biotechnol. 2018, 6, 2. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Jin, J.; Zhang, C.; Yang, X.; Liu, Y.; Lei, P.; Hu, Y. 3D Bioprinting Vascular Networks in Suspension Baths. Appl. Mater. Today 2023, 30, 101729. [Google Scholar] [CrossRef]
- Murphy, S.V.; De Coppi, P.; Atala, A. Opportunities and Challenges of Translational 3D Bioprinting. Nat. Biomed. Eng. 2019, 4, 370–380. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.P.; Toksoy, Z.; Davis, B.A.; Geibel, J.P. 3D Bioprinting of Vascularized Tissues for in Vitro and in Vivo Applications. Front. Bioeng. Biotechnol. 2021, 9, 326. [Google Scholar] [CrossRef] [PubMed]
- Szklanny, A.A.; Machour, M.; Redenski, I.; Chochola, V.; Goldfracht, I.; Kaplan, B.; Epshtein, M.; Simaan Yameen, H.; Merdler, U.; Feinberg, A.; et al. 3D Bioprinting of Engineered Tissue Flaps with Hierarchical Vessel Networks (VesselNet) for Direct Host-To-Implant Perfusion. Adv. Mater. 2021, 33, 2102661. [Google Scholar] [CrossRef] [PubMed]
- Song, K.H.; Highley, C.B.; Rouff, A.; Burdick, J.A. Complex 3D-Printed Microchannels within Cell-Degradable Hydrogels. Adv. Funct. Mater. 2018, 28, 1801331. [Google Scholar] [CrossRef]
- Skylar-Scott, M.A.; Uzel, S.G.M.; Nam, L.L.; Ahrens, J.H.; Truby, R.L.; Damaraju, S.; Lewis, J.A. Biomanufacturing of Organ-Specific Tissues with High Cellular Density and Embedded Vascular Channels. Sci. Adv. 2019, 5, eaaw2459. [Google Scholar] [CrossRef] [PubMed]
- Noor, N.; Shapira, A.; Edri, R.; Gal, I.; Wertheim, L.; Dvir, T. 3D Printing of Personalized Thick and Perfusable Cardiac Patches and Hearts. Adv. Sci. 2019, 6, 1900344. [Google Scholar] [CrossRef]
- Song, M.J.; Quinn, R.; Nguyen, E.; Hampton, C.; Sharma, R.; Park, T.S.; Koster, C.; Voss, T.; Tristan, C.; Weber, C.; et al. Bioprinted 3D Outer Retina Barrier Uncovers RPE-Dependent Choroidal Phenotype in Advanced Macular Degeneration. Nat. Methods 2022, 20, 149–161. [Google Scholar] [CrossRef]
- Schöneberg, J.; De Lorenzi, F.; Theek, B.; Blaeser, A.; Rommel, D.; Kuehne, A.J.C.; Kießling, F.; Fischer, H. Engineering Biofunctional in Vitro Vessel Models Using a Multilayer Bioprinting Technique. Sci. Rep. 2018, 8, 10430. [Google Scholar] [CrossRef]
- Gao, G.; Lee, J.H.; Jang, J.; Lee, D.H.; Kong, J.S.; Kim, B.S.; Choi, Y.J.; Jang, W.B.; Hong, Y.J.; Kwon, S.M.; et al. Tissue Engineered Bio-Blood-Vessels Constructed Using a Tissue-Specific Bioink and 3D Coaxial Cell Printing Technique: A Novel Therapy for Ischemic Disease. Adv. Funct. Mater. 2017, 27, 1700798. [Google Scholar] [CrossRef]
- Silberman, E.; Oved, H.; Namestnikov, M.; Shapira, A.; Dvir, T. Post-Maturation Reinforcement of 3d-Printed Vascularized Cardiac Tissues. Adv. Mater. 2023, 35, 2302229. [Google Scholar] [CrossRef]
- Kolesky, D.B.; Homan, K.A.; Skylar-Scott, M.A.; Lewis, J.A. Three-Dimensional Bioprinting of Thick Vascularized Tissues. Proc. Natl. Acad. Sci. USA 2016, 113, 3179–3184. [Google Scholar] [CrossRef]
- Miller, J.S.; Stevens, K.R.; Yang, M.T.; Baker, B.M.; Nguyen, D.H.T.; Cohen, D.M.; Toro, E.; Chen, A.A.; Galie, P.A.; Yu, X.; et al. Rapid Casting of Patterned Vascular Networks for Perfusable Engineered Three-Dimensional Tissues. Nat. Mater. 2012, 11, 768–774. [Google Scholar] [CrossRef] [PubMed]
- Soffer-Tsur, N.; Shevach, M.; Shapira, A.; Peer, D.; Dvir, T. Optimizing the Biofabrication Process of Omentum-Based Scaffolds for Engineering Autologous Tissues. Biofabrication 2014, 6, 035023. [Google Scholar] [CrossRef] [PubMed]
- Boateng, S.Y.; Lateef, S.S.; Mosley, W.; Hartman, T.J.; Hanley, L.; Russell, B. RGD and YIGSR Synthetic Peptides Facilitate Cellular Adhesion Identical to That of Laminin and Fibronectin but Alter the Physiology of Neonatal Cardiac Myocytes. Am. J. Physiol. Cell Physiol. 2005, 288, C30–C38. [Google Scholar] [CrossRef]
- Shevach, M.; Soffer-Tsur, N.; Fleischer, S.; Shapira, A.; Dvir, T. Fabrication of Omentum-Based Matrix for Engineering Vascularized Cardiac Tissues. Biofabrication 2014, 6, 024101. [Google Scholar] [CrossRef]
- Van Rosmalen, M.; Krom, M.; Merkx, M. Tuning the Flexibility of Glycine-Serine Linkers to Allow Rational Design of Multidomain Proteins. Biochemistry 2017, 56, 6565–6574. [Google Scholar] [CrossRef]
- Kumai, J.; Hozumi, K.; Yamada, Y.; Katagiri, F.; Kikkawa, Y.; Nomizu, M. Effect of Spacer Length and Type on the Biological Activity of Peptide-Polysaccharide Matrices. Pept. Sci. 2016, 106, 512–520. [Google Scholar] [CrossRef]
- Golunova, A.; Velychkivska, N.; Mikšovská, Z.; Chochola, V.; Jaroš, J.; Hampl, A.; Pop-Georgievski, O.; Proks, V. Direct and Indirect Biomimetic Peptide Modification of Alginate: Efficiency, Side Reactions, and Cell Response. Int. J. Mol. Sci. 2021, 22, 5731. [Google Scholar] [CrossRef]
- Shachar, M.; Tsur-Gang, O.; Dvir, T.; Leor, J.; Cohen, S. The Effect of Immobilized RGD Peptide in Alginate Scaffolds on Cardiac Tissue Engineering. Acta Biomater. 2011, 7, 152–162. [Google Scholar] [CrossRef] [PubMed]
- Tsur-Gang, O.; Ruvinov, E.; Landa, N.; Holbova, R.; Feinberg, M.S.; Leor, J.; Cohen, S. The Effects of Peptide-Based Modification of Alginate on Left Ventricular Remodeling and Function after Myocardial Infarction. Biomaterials 2009, 30, 189–195. [Google Scholar] [CrossRef]
- Lien, S.M.; Ko, L.Y.; Huang, T.J. Effect of Pore Size on ECM Secretion and Cell Growth in Gelatin Scaffold for Articular Cartilage Tissue Engineering. Acta Biomater. 2009, 5, 670–679. [Google Scholar] [CrossRef]
- Gerecht, S.; Townsend, S.A.; Pressler, H.; Zhu, H.; Nijst, C.L.E.; Bruggeman, J.P.; Nichol, J.W.; Langer, R. A Porous Photocurable Elastomer for Cell Encapsulation and Culture. Biomaterials 2007, 28, 4826–4835. [Google Scholar] [CrossRef] [PubMed]
- Mandal, B.B.; Kundu, S.C. Cell Proliferation and Migration in Silk Fibroin 3D Scaffolds. Biomaterials 2009, 30, 2956–2965. [Google Scholar] [CrossRef] [PubMed]
- Ehrbar, M.; Sala, A.; Lienemann, P.; Ranga, A.; Mosiewicz, K.; Bittermann, A.; Rizzi, S.C.; Weber, F.E.; Lutolf, M.P. Elucidating the Role of Matrix Stiffness in 3D Cell Migration and Remodeling. Biophys. J. 2011, 100, 284. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Zhao, W.; Xiahou, Z.; Wang, X.; Zhang, K.; Yin, J. Bioink Design for Extrusion-Based Bioprinting. Appl. Mater. Today 2021, 25, 101227. [Google Scholar] [CrossRef]
- Crupi, A.; Costa, A.; Tarnok, A.; Melzer, S.; Teodori, L. Inflammation in Tissue Engineering: The Janus between Engraftment and Rejection. Eur. J. Immunol. 2015, 45, 3222–3236. [Google Scholar] [CrossRef]
- Mansbridge, J. Commercial Considerations in Tissue Engineering. J. Anat. 2006, 209, 527–532. [Google Scholar] [CrossRef]
- Lim, R.G.; Quan, C.; Reyes-Ortiz, A.M.; Lutz, S.E.; Kedaigle, A.J.; Gipson, T.A.; Wu, J.; Vatine, G.D.; Stocksdale, J.; Casale, M.S.; et al. Huntington’s Disease IPSC-Derived Brain Microvascular Endothelial Cells Reveal WNT-Mediated Angiogenic and Blood-Brain Barrier Deficits. Cell Rep. 2017, 19, 1365–1377. [Google Scholar] [CrossRef]
- Grekhnev, D.A.; Kaznacheyeva, E.V.; Vigont, V.A. Patient-Specific IPSCs-Based Models of Neurodegenerative Diseases: Focus on Aberrant Calcium Signaling. Int. J. Mol. Sci. 2022, 23, 624. [Google Scholar] [CrossRef] [PubMed]
- Doulatov, S.; Vo, L.T.; Macari, E.R.; Wahlster, L.; Kinney, M.A.; Taylor, A.M.; Barragan, J.; Gupta, M.; McGrath, K.; Lee, H.Y.; et al. Drug Discovery for Diamond-Blackfan Anemia Using Reprogrammed Hematopoietic Progenitors. Sci. Transl. Med. 2017, 9, eaah5645. [Google Scholar] [CrossRef] [PubMed]
- Qian, L.; Julia, T.C.W. Human IPSC-Based Modeling of Central Nerve System Disorders for Drug Discovery. Int. J. Mol. Sci. 2021, 22, 1203. [Google Scholar] [CrossRef] [PubMed]
- Elitt, M.S.; Barbar, L.; Tesar, P.J. Drug Screening for Human Genetic Diseases Using IPSC Models. Hum. Mol. Genet. 2018, 27, R89–R98. [Google Scholar] [CrossRef] [PubMed]
- Sequiera, G.L.; Srivastava, A.; Sareen, N.; Yan, W.; Alagarsamy, K.N.; Verma, E.; Aghanoori, M.R.; Aliani, M.; Kumar, A.; Fernyhough, P.; et al. Development of IPSC-Based Clinical Trial Selection Platform for Patients with Ultrarare Diseases. Sci. Adv. 2022, 8, 4370. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [PubMed]
- Abutaleb, N.O.; Truskey, G.A. Differentiation and Characterization of Human IPSC-Derived Vascular Endothelial Cells under Physiological Shear Stress. STAR Protoc. 2021, 2, 100394. [Google Scholar] [CrossRef] [PubMed]
- Haudenschild, C.C. Morphology of Vascular Endothelial Cells in Culture. In Biology of Endothelial Cells; Springer: Boston, MA, USA, 1984; pp. 129–140. [Google Scholar] [CrossRef]
- Jiménez, N.; Krouwer, V.J.D.; Post, J.A. A New, Rapid and Reproducible Method to Obtain High Quality Endothelium in Vitro. Cytotechnology 2013, 65, 1. [Google Scholar] [CrossRef]
- Reinhart-King, C.A.; Dembo, M.; Hammer, D.A. The Dynamics and Mechanics of Endothelial Cell Spreading. Biophys. J. 2005, 89, 676–689. [Google Scholar] [CrossRef]
- Cao, X.; Maharjan, S.; Ashfaq, R.; Shin, J.; Zhang, Y.S. Bioprinting of Small-Diameter Blood Vessels. Engineering 2021, 7, 832–844. [Google Scholar] [CrossRef]
- Miri, A.K.; Khalilpour, A.; Cecen, B.; Maharjan, S.; Shin, S.R.; Khademhosseini, A. Multiscale Bioprinting of Vascularized Models. Biomaterials 2019, 198, 204–216. [Google Scholar] [CrossRef] [PubMed]
- Webb, B.; Doyle, B.J. Parameter Optimization for 3D Bioprinting of Hydrogels. Bioprinting 2017, 8, 8–12. [Google Scholar] [CrossRef]
- Mao, A.S.; Shin, J.W.; Mooney, D.J. Effects of Substrate Stiffness and Cell-Cell Contact on Mesenchymal Stem Cell Differentiation. Biomaterials 2016, 98, 184–191. [Google Scholar] [CrossRef]
- Edri, R.; Gal, I.; Noor, N.; Harel, T.; Fleischer, S.; Adadi, N.; Green, O.; Shabat, D.; Heller, L.; Shapira, A.; et al. Personalized Hydrogels for Engineering Diverse Fully Autologous Tissue Implants. Adv. Mater. 2019, 31, 1803895. [Google Scholar] [CrossRef] [PubMed]
- Rowley, J.A.; Madlambayan, G.; Mooney, D.J. Alginate Hydrogels as Synthetic Extracellular Matrix Materials. Biomaterials 1999, 20, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Atchison, L.; Abutaleb, N.O.; Snyder-Mounts, E.; Gete, Y.; Ladha, A.; Ribar, T.; Cao, K.; Truskey, G.A. IPSC-Derived Endothelial Cells Affect Vascular Function in a Tissue-Engineered Blood Vessel Model of Hutchinson-Gilford Progeria Syndrome. Stem Cell Rep. 2020, 14, 325–337. [Google Scholar] [CrossRef] [PubMed]
- Shapira, A.; Noor, N.; Oved, H.; Dvir, T. Transparent Support Media for High Resolution 3D Printing of Volumetric Cell-Containing ECM Structures. Biomed. Mater. 2020, 15, 045018. [Google Scholar] [CrossRef] [PubMed]
- Feyen, D.A.M.; McKeithan, W.L.; Bruyneel, A.A.N.; Spiering, S.; Hörmann, L.; Ulmer, B.; Zhang, H.; Briganti, F.; Schweizer, M.; Hegyi, B.; et al. Metabolic Maturation Media Improve Physiological Function of Human IPSC-Derived Cardiomyocytes. Cell Rep. 2020, 32, 107925. [Google Scholar] [CrossRef]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cohen, R.; Baruch, E.-S.; Cabilly, I.; Shapira, A.; Dvir, T. Modified ECM-Based Bioink for 3D Printing of Multi-Scale Vascular Networks. Gels 2023, 9, 792. https://doi.org/10.3390/gels9100792
Cohen R, Baruch E-S, Cabilly I, Shapira A, Dvir T. Modified ECM-Based Bioink for 3D Printing of Multi-Scale Vascular Networks. Gels. 2023; 9(10):792. https://doi.org/10.3390/gels9100792
Chicago/Turabian StyleCohen, Roni, Ester-Sapir Baruch, Itai Cabilly, Assaf Shapira, and Tal Dvir. 2023. "Modified ECM-Based Bioink for 3D Printing of Multi-Scale Vascular Networks" Gels 9, no. 10: 792. https://doi.org/10.3390/gels9100792