Post-Translational Modification of Proteins Mediated by Nitro-Fatty Acids in Plants: Nitroalkylation
Abstract
:1. Introduction
2. Nitro-Fatty Acids in Animals
3. Nitro-Fatty Acids in Plants
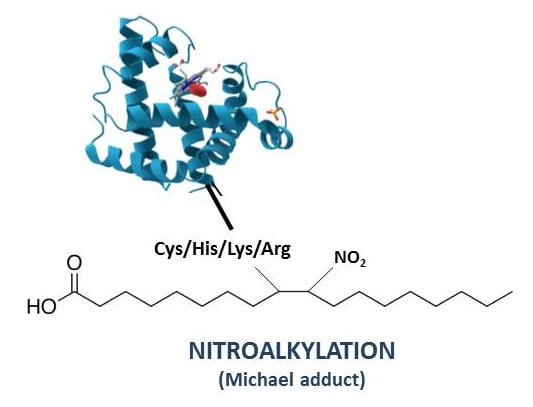
4. Nitroalkylation
4.1. Nitroalkylation in Animals
4.2. Nitroalkylation in Plants
5. Conclusions and Future Perspectives
Funding
Acknowledgments
Conflicts of Interest
References
- Schopfer, F.J.; Cipollina, C.; Freeman, B.A. Formation and signaling actions of electrophilic lipids. Chem. Rev. 2011, 111, 5997–6021. [Google Scholar] [CrossRef] [PubMed]
- Beavers, W.N.; Rose, K.L.; Galligan, J.J.; Mitchener, M.M.; Rouzer, C.A.; Tallman, K.A.; Lamberson, C.R.; Wang, X.; Hill, S.; Ivanova, P.T. Protein modification by endogenously generated lipid electrophiles: Mitochondria as the source and target. ACS Chem. Biol. 2017, 12, 2062–2069. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Weerapana, E.; Blewett, M.M.; Cravatt, B.F. A chemoproteomic platform to quantitatively map targets of lipid-derived electrophiles. Nat. Methods 2013, 11, 79. [Google Scholar] [CrossRef] [PubMed]
- Higdon, A.; Diers, A.R.; Oh, J.Y.; Landar, A.; Darley-Usmar, V.M. Cell signalling by reactive lipid species: New concepts and molecular mechanisms. Biochem. J. 2012, 442, 453–464. [Google Scholar] [CrossRef] [PubMed]
- Marwah, S.; Blann, A.; Rea, C.; Phillips, J.; Wright, J.; Bareford, D. Reduced vitamin E antioxidant capacity in sickle cell disease is related to transfusion status but not to sickle crisis. Am. J. Hematol. 2002, 69, 144–146. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Porter, N.A. New insights regarding the autoxidation of polyunsaturated fatty acids. Antioxid. Redox Signal. 2005, 7, 170–184. [Google Scholar] [CrossRef]
- Poon, H.F.; Calabrese, V.; Scapagnini, G.; Butterfield, D.A. Free radicals: Key to brain aging and heme oxygenase as a cellular response to oxidative stress. J. Gerontol. A Biol. Sci. Med. Sci. 2004, 59, M478–M493. [Google Scholar] [CrossRef]
- Morrow, J.D. Quantification of isoprostanes as indices of oxidant stress and the risk of atherosclerosis in humans. Arterioscler. Thromb. Vasc. Biol. 2005, 25, 279–286. [Google Scholar] [CrossRef]
- Mata-Pérez, C.; Sánchez-Calvo, B.; Padilla, M.N.; Begara-Morales, J.C.; Luque, F.; Melguizo, M.; Jiménez-Ruiz, J.; Fierro-Risco, J.; Peñas-Sanjuán, A.; Valderrama, R. Nitro-fatty acids in plant signaling: Nitro-linolenic acid induces the molecular chaperone network in Arabidopsis. Plant Physiol. 2016, 170, 686–701. [Google Scholar] [CrossRef]
- Mata-Perez, C.; Sanchez-Calvo, B.; Padilla, M.N.; Begara-Morales, J.C.; Valderrama, R.; Corpas, F.J.; Barroso, J.B. Nitro-fatty acids in plant signaling: New key mediators of nitric oxide metabolism. Redox Biol. 2017, 11, 554–561. [Google Scholar] [CrossRef]
- Porter, N.A.; Caldwell, S.E.; Mills, K.A. Mechanisms of free radical oxidation of unsaturated lipids. Lipids 1995, 30, 277–290. [Google Scholar] [CrossRef] [PubMed]
- Addis, P. Occurrence of lipid oxidation products in foods. Food Chem. Toxicol. 1986, 24, 1021–1030. [Google Scholar] [CrossRef]
- Hwa Lee, S.; Rangiah, K.; Williams, M.V.; Wehr, A.Y.; DuBois, R.N.; Blair, I.A. Cyclooxygenase-2-mediated metabolism of arachidonic acid to 15-oxo-eicosatetraenoic acid by rat intestinal epithelial cells. Chem. Res. Toxicol. 2007, 20, 1665–1675. [Google Scholar] [CrossRef]
- Chiang, N.; Serhan, C.N.; Dahlén, S.-E.; Drazen, J.M.; Hay, D.W.; Rovati, G.E.; Shimizu, T.; Yokomizo, T.; Brink, C. The lipoxin receptor ALX: Potent ligand-specific and stereoselective actions in vivo. Pharmacol. Rev. 2006, 58, 463–487. [Google Scholar] [CrossRef] [PubMed]
- Baker, P.R.; Schopfer, F.J.; O’Donnell, V.B.; Freeman, B.A. Convergence of nitric oxide and lipid signaling: Anti-inflammatory nitro-fatty acids. Free Radic. Biol. Med. 2009, 46, 989–1003. [Google Scholar] [CrossRef] [PubMed]
- Rubbo, H.; Radi, R.; Trujillo, M.; Telleri, R.; Kalyanaraman, B.; Barnes, S.; Kirk, M.; Freeman, B.A. Nitric oxide regulation of superoxide and peroxynitrite-dependent lipid peroxidation. Formation of novel nitrogen-containing oxidized lipid derivatives. J. Biol. Chem. 1994, 269, 26066–26075. [Google Scholar] [PubMed]
- Davies, S.S.; Amarnath, V.; Roberts, L.J., II. Isoketals: Highly reactive γ-ketoaldehydes formed from the H2-isoprostane pathway. Chem. Phys. Lipids 2004, 128, 85–99. [Google Scholar] [CrossRef]
- Ueno, N.; Murakami, M.; Tanioka, T.; Fujimori, K.; Tanabe, T.; Urade, Y.; Kudo, I. Coupling between cyclooxygenase, terminal prostanoid synthase, and phospholipase A2. J. Biol. Chem. 2001, 276, 34918–34927. [Google Scholar] [CrossRef]
- Prigge, S.; Boyington, J.; Faig, M.; Doctor, K.; Gaffney, B.; Amzel, L. Structure and mechanism of lipoxygenases. Biochimie 1997, 79, 629–636. [Google Scholar] [CrossRef]
- O’donnell, V.B.; Freeman, B.A. Interactions between nitric oxide and lipid oxidation pathways: Implications for vascular disease. Circ. Res. 2001, 88, 12–21. [Google Scholar] [CrossRef]
- Swain, C.G.; Scott, C.B. Quantitative correlation of relative rates. Comparison of hydroxide ion with other nucleophilic reagents toward alkyl halides, esters, epoxides and acyl halides1. J. Am. Chem. Soc. 1953, 75, 141–147. [Google Scholar] [CrossRef]
- Rudolph, T.K.; Freeman, B.A. Transduction of redox signaling by electrophile-protein reactions. Sci. Signal. 2009, 2, re7. [Google Scholar] [CrossRef] [PubMed]
- Nagahara, N.; Matsumura, T.; Okamoto, R.; Kajihara, Y. Protein cysteine modifications: (2) reactivity specificity and topics of medicinal chemistry and protein engineering. Curr. Med. Chem. 2009, 16, 4490–4501. [Google Scholar] [CrossRef]
- Turell, L.; Steglich, M.; Alvarez, B. The chemical foundations of nitroalkene fatty acid signaling through addition reactions with thiols. Nitric Oxide 2018. [Google Scholar] [CrossRef] [PubMed]
- Pearson, R.G. Hard and soft acids and bases. J. Am. Chem. Soc. 1963, 85, 3533–3539. [Google Scholar] [CrossRef]
- Reed, T.T. Lipid peroxidation and neurodegenerative disease. Free Radic. Biol. Med. 2011, 51, 1302–1319. [Google Scholar] [CrossRef]
- Di Domenico, F.; Pupo, G.; Tramutola, A.; Giorgi, A.; Schininà, M.E.; Coccia, R.; Head, E.; Butterfield, D.A.; Perluigi, M. Redox proteomics analysis of HNE-modified proteins in Down syndrome brain: Clues for understanding the development of Alzheimer disease. Free Radic. Biol. Med. 2014, 71, 270–280. [Google Scholar] [CrossRef]
- Butterfield, D.A.; Gu, L.; Domenico, F.D.; Robinson, R.A. Mass spectrometry and redox proteomics: Applications in disease. Mass Spectrom. Rev. 2014, 33, 277–301. [Google Scholar] [CrossRef]
- Sauriasari, R.; Andrajati, R.; Saputri, D.; Muris, R.; Manfaatun, A.; Amanda, O.; Setiawan, H.; Sakano, N.; Wang, D.; Ogino, K. Marker of lipid peroxidation related to diabetic nephropathy in Indonesian type 2 diabetes mellitus patients. Diabetes Res. Clin. Pract. 2015, 108, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Uchida, K. Role of reactive aldehyde in cardiovascular diseases. Free Radic. Biol. Med. 2000, 28, 1685–1696. [Google Scholar] [CrossRef]
- Bell-Parikh, L.C.; Ide, T.; Lawson, J.A.; McNamara, P.; Reilly, M.; FitzGerald, G.A. Biosynthesis of 15-deoxy-Δ 12, 14-PGJ 2 and the ligation of PPARγ. J. Clin. Investig. 2003, 112, 945–955. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.Y.; Giles, N.; Landar, A.; Darley-Usmar, V. Accumulation of 15-deoxy-Δ12, 14-prostaglandin J2 adduct formation with Keap1 over time: Effects on potency for intracellular antioxidant defence induction. Biochem. J. 2008, 411, 297–306. [Google Scholar] [CrossRef]
- Groeger, A.L.; Cipollina, C.; Cole, M.P.; Woodcock, S.R.; Bonacci, G.; Rudolph, T.K.; Rudolph, V.; Freeman, B.A.; Schopfer, F.J. Cyclooxygenase-2 generates anti-inflammatory mediators from omega-3 fatty acids. Nat. Chem. Biol. 2010, 6, 433–441. [Google Scholar] [CrossRef]
- Khoo, N.K.; Freeman, B.A. Electrophilic nitro-fatty acids: Anti-inflammatory mediators in the vascular compartment. Curr. Opin. Pharmacol. 2010, 10, 179–184. [Google Scholar] [CrossRef]
- Wong, H.L.; Liebler, D.C. Mitochondrial protein targets of thiol-reactive electrophiles. Chem. Res. Toxicol. 2008, 21, 796–804. [Google Scholar] [CrossRef]
- Vila, A.; Tallman, K.A.; Jacobs, A.T.; Liebler, D.C.; Porter, N.A.; Marnett, L.J. Identification of protein targets of 4-hydroxynonenal using click chemistry for ex vivo biotinylation of azido and alkynyl derivatives. Chem. Res. Toxicol. 2008, 21, 432–444. [Google Scholar] [CrossRef]
- Szapacs, M.E.; Kim, H.-Y.H.; Porter, N.A.; Liebler, D.C. Identification of proteins adducted by lipid peroxidation products in plasma and modifications of apolipoprotein A1 with a novel biotinylated phospholipid probe. J. Proteome Res. 2008, 7, 4237–4246. [Google Scholar] [CrossRef]
- Shin, N.-Y.; Liu, Q.; Stamer, S.L.; Liebler, D.C. Protein targets of reactive electrophiles in human liver microsomes. Chem. Res. Toxicol. 2007, 20, 859–867. [Google Scholar] [CrossRef] [PubMed]
- Dennehy, M.K.; Richards, K.A.; Wernke, G.R.; Shyr, Y.; Liebler, D.C. Cytosolic and nuclear protein targets of thiol-reactive electrophiles. Chem. Res. Toxicol. 2006, 19, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Gomi, K.; Yamamoto, H.; Akimitsu, K. Characterization of a lipoxygenase gene in rough lemon induced by Alternaria alternata. J. Gen. Plant Pathol. 2002, 68, 21–30. [Google Scholar] [CrossRef]
- Kim, E.-S.; Choi, E.; Kim, Y.; Cho, K.; Lee, A.; Shim, J.; Rakwal, R.; Agrawal, G.K.; Han, O. Dual positional specificity and expression of non-traditional lipoxygenase induced by wounding and methyl jasmonate in maize seedlings. Plant Mol. Biol. 2003, 52, 1203–1213. [Google Scholar] [CrossRef] [PubMed]
- Montillet, J.-L.; Agnel, J.-P.; Ponchet, M.; Vailleau, F.; Roby, D.; Triantaphylidès, C. Lipoxygenase-mediated production of fatty acid hydroperoxides is a specific signature of the hypersensitive reaction in plants. Plant Physiol. Biochem. 2002, 40, 633–639. [Google Scholar] [CrossRef]
- He, Y.; Fukushige, H.; Hildebrand, D.F.; Gan, S. Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiol. 2002, 128, 876–884. [Google Scholar] [CrossRef] [PubMed]
- Uchida, K. 4-Hydroxy-2-nonenal: A product and mediator of oxidative stress. Prog. Lipid Res. 2003, 42, 318–343. [Google Scholar] [CrossRef]
- Thoma, I.; Loeffler, C.; Sinha, A.K.; Gupta, M.; Krischke, M.; Steffan, B.; Roitsch, T.; Mueller, M.J. Cyclopentenone isoprostanes induced by reactive oxygen species trigger defense gene activation and phytoalexin accumulation in plants. Plant J. 2003, 34, 363–375. [Google Scholar] [CrossRef] [PubMed]
- Geisler, A.C.; Rudolph, T.K. Nitroalkylation—a redox sensitive signaling pathway. BBA-Gen. Subj. 2012, 1820, 777–784. [Google Scholar] [CrossRef] [PubMed]
- Bonacci, G.; Baker, P.R.; Salvatore, S.R.; Shores, D.; Khoo, N.K.; Koenitzer, J.R.; Vitturi, D.A.; Woodcock, S.R.; Golin-Bisello, F.; Watkins, S. Conjugated linoleic acid is a preferential substrate for fatty acid nitration. J. Biol. Chem. 2012, 287, 44071–44082. [Google Scholar] [CrossRef] [PubMed]
- Pryor, W.A.; Lightsey, J.W.; Church, D.F. Reaction of nitrogen dioxide with alkenes and polyunsaturated fatty acids: Addition and hydrogen-abstraction mechanisms. J. Am. Chem. Soc. 1982, 104, 6685–6692. [Google Scholar] [CrossRef]
- Buchan, G.R.; Bonacci, G.; Fazzari, M.; Salvatore, S.; Wendell, S.G. Nitro-fatty acid formation and metabolism. Nitric Oxide 2018, 79, 38–44. [Google Scholar] [CrossRef] [PubMed]
- d’Ischia, M.; Napolitano, A.; Manini, P.; Panzella, L. Secondary targets of nitrite-derived reactive nitrogen species: Nitrosation/nitration pathways, antioxidant defense mechanisms and toxicological implications. Chem. Res. Toxicol. 2011, 24, 2071–2092. [Google Scholar] [CrossRef]
- Rudolph, V.; Rudolph, T.K.; Schopfer, F.J.; Bonacci, G.; Woodcock, S.R.; Cole, M.P.; Baker, P.R.; Ramani, R.; Freeman, B.A. Endogenous generation and protective effects of nitro-fatty acids in a murine model of focal cardiac ischaemia and reperfusion. Cardiovasc. Res. 2009, 85, 155–166. [Google Scholar] [CrossRef] [PubMed]
- Nadtochiy, S.M.; Baker, P.R.; Freeman, B.A.; Brookes, P.S. Mitochondrial nitroalkene formation and mild uncoupling in ischaemic preconditioning: Implications for cardioprotection. Cardiovasc. Res. 2008, 82, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, C.; Roche, H. Conjugated linoleic acid and inflammatory cell signalling. Prostaglandins Leukot. Essent. Fat. Acids 2010, 82, 199–204. [Google Scholar] [CrossRef]
- Vitturi, D.A.; Minarrieta, L.; Salvatore, S.R.; Postlethwait, E.M.; Fazzari, M.; Ferrer-Sueta, G.; Lancaster, J.R., Jr.; Freeman, B.A.; Schopfer, F.J. Convergence of biological nitration and nitrosation via symmetrical nitrous anhydride. Nat. Chem. Biol. 2015, 11, 504. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Yamamoto, M. Stress-sensing mechanisms and the physiological roles of the Keap1-Nrf2 system during cellular stress. J. Biol. Chem. 2017, 292, 16817–16824. [Google Scholar] [CrossRef] [PubMed]
- Weitzberg, E.; Lundberg, J.O. Novel aspects of dietary nitrate and human health. Annu. Rev. Nutr. 2013, 33, 129–159. [Google Scholar] [CrossRef]
- Lundberg, J.O.; Weitzberg, E. NO generation from inorganic nitrate and nitrite: Role in physiology, nutrition and therapeutics. Arch. Pharm. Res. 2009, 32, 1119–1126. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Vivián, C.; Cabello, P.; Martínez-Luque, M.; Blasco, R.; Castillo, F. Prokaryotic nitrate reduction: Molecular properties and functional distinction among bacterial nitrate reductases. J. Bacteriol. 1999, 181, 6573–6584. [Google Scholar]
- Ferreira, A.M.; Ferrari, M.I.; Trostchansky, A.; Batthyany, C.; Souza, J.M.; Alvarez, M.N.; López, G.V.; Baker, P.R.; Schopfer, F.J.; O’Donnell, V. Macrophage activation induces formation of the anti-inflammatory lipid cholesteryl-nitrolinoleate. Biochem. J. 2009, 417, 223–238. [Google Scholar] [CrossRef]
- Freeman, B.A.; Baker, P.R.; Schopfer, F.J.; Woodcock, S.R.; Napolitano, A.; d’Ischia, M. Nitro-fatty acid formation and signaling. J. Biol. Chem. 2008, 283, 15515–15519. [Google Scholar] [CrossRef]
- Faine, L.A.; Cavalcanti, D.; Rudnicki, M.; Ferderbar, S.; Macedo, S.; Souza, H.P.; Farsky, S.; Boscá, L.; Abdalla, D. Bioactivity of nitrolinoleate: Effects on adhesion molecules and CD40-CD40L system. J. Nutr. Biochem. 2010, 21, 125–132. [Google Scholar] [CrossRef]
- Schopfer, F.J.; Baker, P.R.; Giles, G.; Chumley, P.; Batthyany, C.; Crawford, J.; Patel, R.P.; Hogg, N.; Branchaud, B.P.; Lancaster, J.R. Fatty acid transduction of nitric oxide signaling Nitrolinoleic acid is a hydrophobically stabilized nitric oxide donor. J. Biol. Chem. 2005, 280, 19289–19297. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.H.; Wu, S.S.; Hu, C.H. Release of nitric oxide from nitrated fatty acids: Insights from computational chemistry. J. Chin. Chem. Soc. 2019, 66, 41–48. [Google Scholar] [CrossRef]
- Lima, É.S.; Bonini, M.G.; Augusto, O.; Barbeiro, H.V.; Souza, H.P.; Abdalla, D.S. Nitrated lipids decompose to nitric oxide and lipid radicals and cause vasorelaxation. Free Radic. Biol. Med. 2005, 39, 532–539. [Google Scholar] [CrossRef]
- Gorczynski, M.J.; Huang, J.; Lee, H.; King, S.B. Evaluation of nitroalkenes as nitric oxide donors. Bioorg. Med. Chem. Lett. 2007, 17, 2013–2017. [Google Scholar] [CrossRef]
- Batthyany, C.; Schopfer, F.J.; Baker, P.R.; Durán, R.; Baker, L.M.; Huang, Y.; Cerveñansky, C.; Braunchaud, B.P.; Freeman, B.A. Reversible post-translational modification of proteins by nitrated fatty acids in vivo. J. Biol. Chem. 2006, 281, 20450–20463. [Google Scholar] [CrossRef]
- Baker, L.M.; Baker, P.R.; Golin-Bisello, F.; Schopfer, F.J.; Fink, M.; Woodcock, S.R.; Branchaud, B.P.; Radi, R.; Freeman, B.A. Nitro-fatty acid reaction with glutathione and cysteine Kinetic analysis of thiol alkylation by a Michael addition reaction. J. Biol. Chem. 2007, 282, 31085–31093. [Google Scholar] [CrossRef] [PubMed]
- Rubbo, H.; Radi, R. Protein and lipid nitration: Role in redox signaling and injury. BBA-Gen. Subj. 2008, 1780, 1318–1324. [Google Scholar] [CrossRef]
- Fazzari, M.; Khoo, N.; Woodcock, S.R.; Li, L.; Freeman, B.A.; Schopfer, F.J. Generation and esterification of electrophilic fatty acid nitroalkenes in triacylglycerides. Free Radic. Biol. Med. 2015, 87, 113–124. [Google Scholar] [CrossRef]
- Fazzari, M.; Trostchansky, A.; Schopfer, F.J.; Salvatore, S.R.; Sánchez-Calvo, B.; Vitturi, D.; Valderrama, R.; Barroso, J.B.; Radi, R.; Freeman, B.A. Olives and olive oil are sources of electrophilic fatty acid nitroalkenes. PLoS ONE 2014, 9, e84884. [Google Scholar] [CrossRef]
- Vitturi, D.A.; Chen, C.-S.; Woodcock, S.R.; Salvatore, S.R.; Bonacci, G.; Koenitzer, J.R.; Stewart, N.A.; Wakabayashi, N.; Kensler, T.W.; Freeman, B.A. Modulation of nitro-fatty acid signaling prostaglandin reductase-1 is a nitroalkene reductase. J. Biol. Chem. 2013, 288, 25626–25637. [Google Scholar] [CrossRef] [PubMed]
- Melo, T.N.; Domingues, P.; Ferreira, R.; Milic, I.; Fedorova, M.; Santos, S.R.M.; Segundo, M.A.; Domingues, M.R.r.M. Recent advances on mass spectrometry analysis of nitrated phospholipids. Anal. Chem. 2016, 88, 2622–2629. [Google Scholar] [CrossRef] [PubMed]
- Catharino, R.R.; Haddad, R.; Cabrini, L.G.; Cunha, I.B.; Sawaya, A.C.; Eberlin, M.N. Characterization of vegetable oils by electrospray ionization mass spectrometry fingerprinting: Classification, quality, adulteration, and aging. Anal. Chem. 2005, 77, 7429–7433. [Google Scholar] [CrossRef]
- Rastrelli, L.; Passi, S.; Ippolito, F.; Vacca, G.; De Simone, F. Rate of degradation of α-tocopherol, squalene, phenolics, and polyunsaturated fatty acids in olive oil during different storage conditions. J. Agric. Food Chem. 2002, 50, 5566–5570. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Camino, M.C.; Moreda, W.; Mateos, R.; Cert, A. Determination of esters of fatty acids with low molecular weight alcohols in olive oils. J. Agric. Food Chem. 2002, 50, 4721–4725. [Google Scholar] [CrossRef]
- Mata-Pérez, C.; Sánchez-Calvo, B.; Begara-Morales, J.C.; Carreras, A.; Padilla, M.N.; Melguizo, M.; Valderrama, R.; Corpas, F.J.; Barroso, J.B. Nitro-linolenic acid is a nitric oxide donor. Nitric Oxide 2016, 57, 57–63. [Google Scholar] [CrossRef]
- Beligni, M.V.; Lamattina, L. Nitric oxide stimulates seed germination and de-etiolation, and inhibits hypocotyl elongation, three light-inducible responses in plants. Planta 2000, 210, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Bethke, P.C.; Libourel, I.G.; Jones, R.L. Nitric oxide reduces seed dormancy in Arabidopsis. J. Exp. Bot. 2005, 57, 517–526. [Google Scholar] [CrossRef]
- Libourel, I.G.; Bethke, P.C.; De Michele, R.; Jones, R.L. Nitric oxide gas stimulates germination of dormant Arabidopsis seeds: Use of a flow-through apparatus for delivery of nitric oxide. Planta 2006, 223, 813–820. [Google Scholar] [CrossRef] [PubMed]
- Mata-Pérez, C.; Padilla, M.N.; Sánchez-Calvo, B.; Begara-Morales, J.C.; Valderrama, R.; Chaki, M.; Barroso, J.B. Biological properties of nitro-fatty acids in plants. Nitric Oxide 2018. [Google Scholar] [CrossRef] [PubMed]
- Padilla, M.N.; Mata-Pérez, C.; Melguizo, M.; Barroso, J.B. In vitro nitro-fatty acid release from Cys-NO2-fatty acid adducts under nitro-oxidative conditions. Nitric Oxide 2017, 68, 14–22. [Google Scholar] [CrossRef] [PubMed]
- Kansanen, E.; Jyrkkanen, H.-K.; Volger, O.L.; Leinonen, H.; Kivela, A.M.; Hakkinen, S.-K.; Woodcock, S.R.; Schopfer, F.J.; Horrevoets, A.J.; Yla-Herttala, S. Nrf2-dependent and-independent responses to nitro-fatty acids in human endothelial cells: Identification of heat shock response as the major pathway activated by nitro-oleic acid. J. Biol. Chem. 2009, 284, 33233–33241. [Google Scholar] [CrossRef]
- Nishizawa, A.; Yabuta, Y.; Yoshida, E.; Maruta, T.; Yoshimura, K.; Shigeoka, S. Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress. Plant J. 2006, 48, 535–547. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Calvo, B.; Barroso, J.B.; Corpas, F.J. Hypothesis: Nitro-fatty acids play a role in plant metabolism. Plant Sci. 2013, 199, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Astier, J.; Kulik, A.; Koen, E.; Besson-Bard, A.; Bourque, S.; Jeandroz, S.; Lamotte, O.; Wendehenne, D. Protein S-nitrosylation: What’s going on in plants? Free Radic. Biol. Med. 2012, 53, 1101–1110. [Google Scholar] [CrossRef] [PubMed]
- Begara-Morales, J.C.; Chaki, M.; Sánchez-Calvo, B.; Mata-Pérez, C.; Leterrier, M.; Palma, J.M.; Barroso, J.B.; Corpas, F.J. Protein tyrosine nitration in pea roots during development and senescence. J. Exp. Bot. 2013, 64, 1121–1134. [Google Scholar] [CrossRef] [PubMed]
- Chaki, M.; Shekariesfahlan, A.; Ageeva, A.; Mengel, A.; von Toerne, C.; Durner, J.; Lindermayr, C. Identification of nuclear target proteins for S-nitrosylation in pathogen-treated Arabidopsis thaliana cell cultures. Plant Sci. 2015, 238, 115–126. [Google Scholar] [CrossRef]
- Rudolph, V.; Schopfer, F.J.; Khoo, N.K.; Rudolph, T.K.; Cole, M.P.; Woodcock, S.R.; Bonacci, G.; Groeger, A.L.; Golin-Bisello, F.; Chen, C.-S. Nitro-fatty acid metabolome: Saturation, desaturation, β-oxidation, and protein adduction. J. Biol. Chem. 2009, 284, 1461–1473. [Google Scholar] [CrossRef]
- Melo, T.; Montero-Bullón, J.-F.; Domingues, P.; Domingues, M.R. Discovery of bioactive nitrated lipids and nitro-lipid-protein adducts using mass spectrometry-based approaches. Redox Biol. 2019, 2019, 101106. [Google Scholar] [CrossRef]
- Lin, W.-W.; Jang, Y.-J.; Wang, Y.; Liu, J.-T.; Hu, S.-R.; Wang, L.-Y.; Yao, C.-F. An improved and easy method for the preparation of 2, 2-disubstituted 1-nitroalkenes. J. Org. Chem. 2001, 66, 1984–1991. [Google Scholar] [CrossRef]
- Jang, Y.-J.; Lin, W.-W.; Shih, Y.-K.; Liu, J.-T.; Hwang, M.-H.; Yao, C.-F. A one-pot, two step synthesis of 2, 2-disubstituted 1-nitroalkenes. Tetrahedron 2003, 59, 4979–4992. [Google Scholar] [CrossRef]
- Sawa, T.; Arimoto, H.; Akaike, T. Regulation of redox signaling involving chemical conjugation of protein thiols by nitric oxide and electrophiles. Bioconjugate Chem. 2010, 21, 1121–1129. [Google Scholar] [CrossRef]
- Winterbourn, C.C.; Hampton, M.B. Thiol chemistry and specificity in redox signaling. Free Radic. Biol. Med. 2008, 45, 549–561. [Google Scholar] [CrossRef] [PubMed]
- Trostchansky, A.; Rubbo, H. Nitrated fatty acids: Mechanisms of formation, chemical characterization, and biological properties. Free Radic. Biol. Med. 2008, 44, 1887–1896. [Google Scholar] [CrossRef] [PubMed]
- Delmastro-Greenwood, M.; Freeman, B.A.; Wendell, S.G. Redox-dependent anti-inflammatory signaling actions of unsaturated fatty acids. Annu. Rev. Physiol. 2014, 76, 79–105. [Google Scholar] [CrossRef] [PubMed]
- Woodcock, C.-S.C.; Huang, Y.; Woodcock, S.R.; Salvatore, S.R.; Singh, B.; Golin-Bisello, F.; Davidson, N.E.; Neumann, C.A.; Freeman, B.A.; Wendell, S.G. Nitro-fatty acid inhibition of triple-negative breast cancer cell viability, migration, invasion, and tumor growth. J. Biol. Chem. 2018, 293, 1120–1137. [Google Scholar] [CrossRef]
- Villacorta, L.; Zhang, J.; Garcia-Barrio, M.T.; Chen, X.-l.; Freeman, B.A.; Chen, Y.E.; Cui, T. Nitro-linoleic acid inhibits vascular smooth muscle cell proliferation via the Keap1/Nrf2 signaling pathway. Am. J. Physiol. Heart Circ. Physiol. 2007, 293, H770–H776. [Google Scholar] [CrossRef]
- Cui, T.; Schopfer, F.J.; Zhang, J.; Chen, K.; Ichikawa, T.; Baker, P.R.; Batthyany, C.; Chacko, B.K.; Feng, X.; Patel, R.P. Nitrated fatty acids: Endogenous anti-inflammatory signaling mediators. J. Biol. Chem. 2006, 281, 35686–35698. [Google Scholar] [CrossRef]
- Ambrozova, G.; Martiskova, H.; Koudelka, A.; Ravekes, T.; Rudolph, T.K.; Klinke, A.; Rudolph, V.; Freeman, B.A.; Woodcock, S.R.; Kubala, L. Nitro-oleic acid modulates classical and regulatory activation of macrophages and their involvement in pro-fibrotic responses. Free Radic. Biol. Med. 2016, 90, 252–260. [Google Scholar] [CrossRef]
- Ambrozova, G.; Fidlerova, T.; Verescakova, H.; Koudelka, A.; Rudolph, T.K.; Woodcock, S.R.; Freeman, B.A.; Kubala, L.; Pekarova, M. Nitro-oleic acid inhibits vascular endothelial inflammatory responses and the endothelial-mesenchymal transition. BBA-Gen. Subj. 2016, 1860, 2428–2437. [Google Scholar] [CrossRef]
- Rom, O.; Khoo, N.K.; Chen, Y.E.; Villacorta, L. Inflammatory signaling and metabolic regulation by nitro-fatty acids. Nitric Oxide 2018, 78, 140–145. [Google Scholar] [CrossRef] [PubMed]
- Deen, A.J.; Sihvola, V.; Härkönen, J.; Patinen, T.; Adinolfi, S.; Levonen, A.-L. Regulation of stress signaling pathways by nitro-fatty acids. Nitric Oxide 2018, 78, 170–175. [Google Scholar] [CrossRef] [PubMed]
- Kansanen, E.; Bonacci, G.; Schopfer, F.J.; Kuosmanen, S.M.; Tong, K.I.; Leinonen, H.; Woodcock, S.R.; Yamamoto, M.; Carlberg, C.; Ylä-Herttuala, S. Electrophilic nitro-fatty acids activate NRF2 by a KEAP1 cysteine 151-independent mechanism. J. Biol. Chem. 2011, 286, 14019–14027. [Google Scholar] [CrossRef]
- Dinkova-Kostova, A.T.; Holtzclaw, W.D.; Cole, R.N.; Itoh, K.; Wakabayashi, N.; Katoh, Y.; Yamamoto, M.; Talalay, P. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc. Natl. Acad. Sci. USA 2002, 99, 11908–11913. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Amarilla, P.; Miquel, E.; Trostchansky, A.; Trias, E.; Ferreira, A.M.; Freeman, B.A.; Cassina, P.; Barbeito, L.; Vargas, M.R.; Rubbo, H. Electrophilic nitro-fatty acids prevent astrocyte-mediated toxicity to motor neurons in a cell model of familial amyotrophic lateral sclerosis via nuclear factor erythroid 2-related factor activation. Free Radic. Biol. Med. 2016, 95, 112–120. [Google Scholar] [CrossRef] [PubMed]
- Benjamin, I.J.; McMillan, D.R. Stress (heat shock) proteins: Molecular chaperones in cardiovascular biology and disease. Circ. Res. 1998, 83, 117–132. [Google Scholar] [CrossRef] [PubMed]
- Vihervaara, A.; Sistonen, L. HSF1 at a glance. J. Cell Sci. 2014, 127, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, J.; Schopfer, F.J.; Martynowski, D.; Garcia-Barrio, M.T.; Kovach, A.; Suino-Powell, K.; Baker, P.R.; Freeman, B.A.; Chen, Y.E. Molecular recognition of nitrated fatty acids by PPARγ. Nat. Struct. Mol. Biol. 2008, 15, 865. [Google Scholar] [CrossRef]
- Baker, P.; Schopfer, F.; Batthyany, C.; Groeger, A.; Branchaud, B.; Chen, Y.; Freeman, B. multiple Nitrated Unsaturated Fatty Acid Derivatives Exist In Human Blood And Urine And Serve As Endogenous Ppar Ligands: 261. Free Radic. Biol. Med. 2005, 39, S97. [Google Scholar]
- Schopfer, F.J.; Cole, M.P.; Groeger, A.L.; Chen, C.-S.; Khoo, N.K.; Woodcock, S.R.; Golin-Bisello, F.; Motanya, U.N.; Li, Y.; Zhang, J. Covalent peroxisome proliferator-activated receptor γ adduction by nitro-fatty acids selective ligand activity and anti-diabetic signaling actions. J. Biol. Chem. 2010, 285, 12321–12333. [Google Scholar] [CrossRef] [PubMed]
- Villacorta, L.; Minarrieta, L.; Salvatore, S.R.; Khoo, N.K.; Rom, O.; Gao, Z.; Berman, R.C.; Jobbagy, S.; Li, L.; Woodcock, S.R. In situ generation, metabolism and immunomodulatory signaling actions of nitro-conjugated linoleic acid in a murine model of inflammation. Redox Biol. 2018, 15, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Khoo, N.K.; Li, L.; Salvatore, S.R.; Schopfer, F.J.; Freeman, B.A. Electrophilic fatty acid nitroalkenes regulate Nrf2 and NF-κB signaling: A medicinal chemistry investigation of structure-function relationships. Sci. Rep. 2018, 8, 2295. [Google Scholar] [CrossRef] [PubMed]
- Villacorta, L.; Chang, L.; Salvatore, S.R.; Ichikawa, T.; Zhang, J.; Petrovic-Djergovic, D.; Jia, L.; Carlsen, H.; Schopfer, F.J.; Freeman, B.A. Electrophilic nitro-fatty acids inhibit vascular inflammation by disrupting LPS-dependent TLR4 signalling in lipid rafts. Cardiovasc. Res. 2013, 98, 116–124. [Google Scholar] [CrossRef]
- Ricote, M.; Glass, C.K. PPARs and molecular mechanisms of transrepression. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2007, 1771, 926–935. [Google Scholar] [CrossRef] [PubMed]
- Bonacci, G.; Schopfer, F.J.; Batthyany, C.I.; Rudolph, T.K.; Rudolph, V.; Khoo, N.K.; Kelley, E.E.; Freeman, B.A. Electrophilic fatty acids regulate matrix metalloproteinase activity and expression. J. Biol. Chem. 2011, 286, 16074–16081. [Google Scholar] [CrossRef] [PubMed]
- Artim, D.; Bazely, F.; Daugherty, S.; Sculptoreanu, A.; Koronowski, K.; Schopfer, F.; Woodcock, S.; Freeman, B.; de Groat, W. Nitro-oleic acid targets transient receptor potential (TRP) channels in capsaicin sensitive afferent nerves of rat urinary bladder. Exp. Neurol. 2011, 232, 90–99. [Google Scholar] [CrossRef]
- Sculptoreanu, A.; Kullmann, F.; Artim, D.; Bazley, F.; Schopfer, F.; Woodcock, S.; Freeman, B.; De Groat, W. Nitro-oleic acid inhibits firing and activates TRPV1-and TRPA1-mediated inward currents in dorsal root ganglion neurons from adult male rats. J. Pharmacol. Exp. Ther. 2010, 333, 883–895. [Google Scholar] [CrossRef]
- Zhang, J.; Villacorta, L.; Chang, L.; Fan, Z.; Hamblin, M.; Zhu, T.; Chen, C.S.; Cole, M.P.; Schopfer, F.J.; Deng, C.X. Nitro-oleic acid inhibits angiotensin II–induced hypertension. Circ. Res. 2010, 107, 540–548. [Google Scholar] [CrossRef] [PubMed]
- Gil, M.; Graña, M.; Schopfer, F.J.; Wagner, T.; Denicola, A.; Freeman, B.A.; Alzari, P.M.; Batthyány, C.; Durán, R. Inhibition of Mycobacterium tuberculosis PknG by non-catalytic rubredoxin domain specific modification: Reaction of an electrophilic nitro-fatty acid with the Fe–S center. Free Radic. Biol. Med. 2013, 65, 150–161. [Google Scholar] [CrossRef]
- Kelley, E.E.; Batthyany, C.I.; Hundley, N.J.; Woodcock, S.R.; Bonacci, G.; Del Rio, J.M.; Schopfer, F.J.; Lancaster, J.R.; Freeman, B.A.; Tarpey, M.M. Nitro-oleic acid, a novel and irreversible inhibitor of xanthine oxidoreductase. J. Biol. Chem. 2008, 283, 36176–36184. [Google Scholar] [CrossRef]
- Nadtochiy, S.M.; Zhu, Q.; Urciuoli, W.; Rafikov, R.; Black, S.M.; Brookes, P.S. Nitroalkenes confer acute cardioprotection via adenine nucleotide translocase 1. J. Biol. Chem. 2012, 287, 3573–3580. [Google Scholar] [CrossRef] [PubMed]
- Turell, L.; Vitturi, D.A.; Coitiño, E.L.; Lebrato, L.; Möller, M.N.; Sagasti, C.; Salvatore, S.R.; Woodcock, S.R.; Alvarez, B.; Schopfer, F.J. The chemical basis of thiol addition to nitro-conjugated linoleic acid, a protective cell-signaling lipid. J. Biol. Chem. 2017, 292, 1145–1159. [Google Scholar] [CrossRef] [PubMed]
- Trostchansky, A.; Bonilla, L.; Thomas, C.P.; O’Donnell, V.B.; Marnett, L.J.; Radi, R.; Rubbo, H. Nitroarachidonic acid, a novel peroxidase inhibitor of prostaglandin endoperoxide H synthases 1 and 2. J. Biol. Chem. 2011, 286, 12891–12900. [Google Scholar] [CrossRef] [PubMed]
- Bonilla, L.; O‘Donnell, V.; Clark, S.; Rubbo, H.; Trostchansky, A. Regulation of protein kinase C by nitroarachidonic acid: Impact on human platelet activation. Arch. Biochem. Biophys. 2013, 533, 55–61. [Google Scholar] [CrossRef]
- González-Perilli, L.; Álvarez, M.N.; Prolo, C.; Radi, R.; Rubbo, H.; Trostchansky, A. Nitroarachidonic acid prevents NADPH oxidase assembly and superoxide radical production in activated macrophages. Free Radic. Biol. Med. 2013, 58, 126–133. [Google Scholar] [CrossRef] [PubMed]
- González-Perilli, L.; Mastrogiovanni, M.; de Castro Fernandes, D.; Rubbo, H.; Laurindo, F.; Trostchansky, A. Nitroarachidonic acid (NO2AA) inhibits protein disulfide isomerase (PDI) through reversible covalent adduct formation with critical cysteines. BBA-Gen. Subj. 2017, 1861, 1131–1139. [Google Scholar] [CrossRef]
- Asada, K. Ascorbate peroxidase—A hydrogen peroxide-scavenging enzyme in plants. Physiol. Plant. 1992, 85, 235–241. [Google Scholar] [CrossRef]
- Noctor, G.; Foyer, C.H. Ascorbate and glutathione: Keeping active oxygen under control. Annu. Rev. Plant Biol. 1998, 49, 249–279. [Google Scholar] [CrossRef]
- Hossain, M.A.; Asada, K. Inactivation of ascorbate peroxidase in spinach chloroplasts on dark addition of hydrogen peroxide: Its protection by ascorbate. Plant Cell Physiol. 1984, 25, 1285–1295. [Google Scholar]
- Metcalfe, C.; Macdonald, I.K.; Murphy, E.J.; Brown, K.A.; Raven, E.L.; Moody, P.C. The tuberculosis prodrug isoniazid bound to activating peroxidases. J. Biol. Chem. 2008, 283, 6193–6200. [Google Scholar] [CrossRef]








| Name | Formula | Chemical Structure |
|---|---|---|
| Nitro-oleic acid (9-, and 10-nitro-all-cis-octadecaenoic acid) | NO2-OA (18:1) |  |
| Nitro-linoleic acid (9-, 10-, 12-, and 13-nitro-all-cis octadecadienoic acid) | NO2-LA (18:2) |  |
| Nitro-linolenic acid (9-, 10-, 12-, 13-, 15- and 16-nitro-all-cis-octadecatrienoic acid) | NO2-Ln (18:3) |  |
| Nitro-arachidonic acid (5-, 6-, 8-, 9-, 11-, 12-, 14- and 15-nitro-all-cis-eicosatetraenoic acid) | NO2-AA (20:4) |  |
| Nitro-Fatty Acid | Protein | Nucleophile Site | Effect | References |
|---|---|---|---|---|
| NO2-OA | GAPDH | Catalytic Cys, other Cys and His | Inhibition, increase in hydrophobicity and change in subcellular distribution | [66] |
| Pro-MMP7 and Pro-MMP9 | Zinc coordination Cys in the active site | Zinc release, autocatalytic cleavage of the pro-domain. MMP activation | [115] | |
| TRPV1 and TRPA1 | Not detected | Activation of TRP channels | [116,117] | |
| AT1R | Not detected | Decrease in coupling with G-protein, inhibition of downstream signaling | [118] | |
| PknG | Iron coordination Cys in non-catalytic domain and His | Inhibition of kinase activity | [119] | |
| XOR | Pterindithiolene which coordinates molybdenum | Inhibition of electron transfer reactions at the molybdenum cofactor | [120] | |
| HSF1 | Not detected | Activation of HSFA1 and subsequent robust induction of heat shock genes | [82,107] | |
| NO2-LA | ANT1 | Cys | Cardio-protection | [121] |
| NO2-cLA | HSA | Cys and non-covalent binding | [122] | |
| NO2-AA | PGHS | Disruption of heme binding to the protein | Inhibition of PGHS-1 cyclooxygenase activity and both PGHS-1 and -2 peroxidase activity | [123] |
| PKC | Probable covalent modification | Inhibitory effect on PKC activation | [124] | |
| NOX2 | Inhibition of assembly | Inhibition of superoxide production | [125] | |
| PDI | Cys at active site | Inhibition of reductase and chaperone activities | [126] | |
| NO2-OA and NO2-LA | NF-κB p65 | DNA binding domain Cys | Inhibition of NF-κB DNA binding, abolition of pro-inflammatory responses | [98] |
| PPARγ | Cys in ligand-binding domain | Agonist activation of PPARγ | [110] | |
| NO2-OA, NO2-LA and NO2-AA | Keap 1 | Cys | Stabilization of the complex with Nrf2, newly synthesized Nrf2 translocated to the nucleus | [97,103,104,105] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aranda-Caño, L.; Sánchez-Calvo, B.; Begara-Morales, J.C.; Chaki, M.; Mata-Pérez, C.; Padilla, M.N.; Valderrama, R.; Barroso, J.B. Post-Translational Modification of Proteins Mediated by Nitro-Fatty Acids in Plants: Nitroalkylation. Plants 2019, 8, 82. https://doi.org/10.3390/plants8040082
Aranda-Caño L, Sánchez-Calvo B, Begara-Morales JC, Chaki M, Mata-Pérez C, Padilla MN, Valderrama R, Barroso JB. Post-Translational Modification of Proteins Mediated by Nitro-Fatty Acids in Plants: Nitroalkylation. Plants. 2019; 8(4):82. https://doi.org/10.3390/plants8040082
Chicago/Turabian StyleAranda-Caño, Lorena, Beatriz Sánchez-Calvo, Juan C. Begara-Morales, Mounira Chaki, Capilla Mata-Pérez, María N. Padilla, Raquel Valderrama, and Juan B. Barroso. 2019. "Post-Translational Modification of Proteins Mediated by Nitro-Fatty Acids in Plants: Nitroalkylation" Plants 8, no. 4: 82. https://doi.org/10.3390/plants8040082