Berry Anthocyanin, Acid, and Volatile Trait Analyses in a Grapevine-Interspecific F2 Population Using an Integrated GBS and rhAmpSeq Genetic Map
Abstract
:1. Introduction
2. Results
2.1. Segregation Distortion Analysis and Distortion Threshold Estimation for VRS-F2 Population
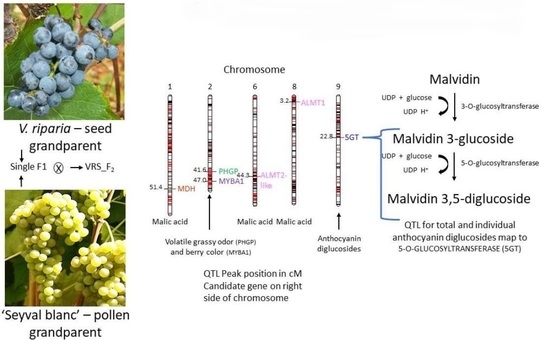
2.2. Map Validation with Berry Skin Color and Flower Type
2.3. Berry Anthocyanin, Acid, and Volatile Analyses
3. Discussion
3.1. Natural Segregation Distortion Was Significant in F2 Population
3.2. The VRS-F2 GBS-rhAmpSeq Integrated Map Provides Greater Marker Transferability to Other Populations
3.3. Confirmation of Berry Skin Color and Flower Type Loci
3.4. Individual Diglucoside Anthocyanin QTL Colocate
3.5. QTL Mapping Identified Malic Acid Dehydrogenase Candidate Gene on Chromosome 1 and Suggests Potential Temperature Influence
3.6. Berry Volatiles and (E)-2-Hexenal QTL
4. Materials and Methods
4.1. Plant Materials
4.2. VRS-F2 GBS-rhAmpSeq Integrated Genetic Map Construction
4.3. Flower, Fruit, and Temperature Data
4.4. Berry Titrable Acidity, Malic Acid, Anthocyanins, and Volatile Measurements
4.5. Statistical Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Töpfer, R.; Hausmann, L.; Harst, M.; Maul, E.; Zyprian, E.; Eibach, R. New horizons for grapevine breeding. Methods Temp. Fruit Breed. 2011, 5, 79–100. [Google Scholar]
- Dalla Costa, L.; Malnoy, M.; Gribaudo, I. Breeding next generation tree fruits: Technical and legal challenges. Horti. Res. 2017, 4, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kambiranda, D.; Obuya, J.; Snowden, J. Grapevine Improvement through Biotechnology. In Genetic Transformation in Crops; IntechOpen: London, UK, 2020. [Google Scholar]
- He, J.; Zhao, X.; Laroche, A.; Lu, Z.-X.; Liu, H.; Li, Z. Genotyping-by-sequencing (GBS), an ultimate marker-assisted selection (MAS) tool to accelerate plant breeding. Front. Plant Sci. 2014, 5, 484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hyma, K.E.; Barba, P.; Wang, M.; Londo, J.P.; Acharya, C.B.; Mitchell, S.E.; Sun, Q.; Reisch, B.; Cadle-Davidson, L. Hetero-zygous mapping strategy (HetMappS) for high resolution genotyping-by-sequencing markers: A case study in grapevine. PLoS ONE 2015, 10, 4880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zou, C.; Karn, A.; Reisch, B.; Nguyen, A.; Sun, Y.; Bao, Y.; Campbell, M.S.; Church, D.; Williams, S.; Xu, X. Haplotyping the Vitis collinear core genome with rhAmpSeq improves marker transferability in a diverse genus. Nat. Commun. 2020, 11, 413. [Google Scholar] [CrossRef] [Green Version]
- Buckler, E.S.; Ilut, D.C.; Wang, X.; Kretzschmar, T.; Gore, M.; Mitchell, S.E. rAmpSeq: Using repetitive sequences for robust genotyping. bioRxiv 2016, 096628. [Google Scholar] [CrossRef]
- Doligez, A.; Bouquet, A.; Danglot, Y.; Lahogue, F.; Riaz, S.; Meredith, C.; Edwards, K.; This, P. Genetic mapping of grapevine (Vitis vinifera L.) applied to the detection of QTLs for seedlessness and berry weight. Theor. Appl. Genet. 2002, 105, 780–795. [Google Scholar] [CrossRef]
- Kobayashi, S.; Goto-Yamamoto, N.; Hirochika, H. Retrotransposon-induced mutations in grape skin color. Science 2004, 304, 982. [Google Scholar] [CrossRef]
- Fournier-Level, A.; le Cunff, L.; Gomez, C.; Doligez, A.; Ageorges, A.; Roux, C.; Bertrand, Y.; Souquet, J.-M.; Cheynier, V.; This, P. Quantitative genetic bases of anthocyanin variation in grape (Vitis vinifera L. ssp. sativa) berry: A quantitative trait lo-cus to quantitative trait nucleotide integrated study. Genetics 2009, 183, 1127–1139. [Google Scholar] [CrossRef] [Green Version]
- Hausmann, L.; Maul, E.; Ganesch, A.; Töpfer, R. Overview of Genetic Loci for Traits in Grapevine and Their Inte-Gration into the VIVC Database. In Proceedings of the XII International Conference on Grapevine Breeding and Genetics, Bordeaux, France, 30 August 2019; pp. 221–226. [Google Scholar]
- Guo, D.-L.; Zhao, H.-L.; Li, Q.; Zhang, G.-H.; Jiang, J.-F.; Liu, C.-H.; Yu, Y.-H. Genome-wide association study of berry-related traits in grape [Vitis vinifera L.] based on genotyping-by-sequencing markers. Hortic. Res. 2019, 6, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Röckel, F.; Moock, C.; Braun, U.; Schwander, F.; Cousins, P.; Maul, E.; Töpfer, R.; Hausmann, L. Color intensity of the red-fleshed berry phenotype of Vitis vinifera teinturier grapes varies due to a 408 bp duplication in the promoter of VvmybA1. Genes 2020, 11, 891. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Li, S.; Jiang, J.; Tang, X.; Fan, X.; Zhang, Y.; Liu, J.; Liu, C. New quantitative trait locus (QTLs) and candidate genes associated with the grape berry color trait identified based on a high-density genetic map. BMC Plant Biol. 2020, 20, 302. [Google Scholar] [CrossRef] [PubMed]
- Viana, A.; Riaz, S.; Walker, M. Genetic dissection of agronomic traits within a segregating population of breeding table grapes. Genet. Mol. Res. 2013, 12, 951–964. [Google Scholar] [CrossRef] [PubMed]
- Burtch, C.; Mansfield, A.K. Comparing Red Wine Color in V. Vinifera and Hybrid Cultivars. Appellation Cornell Research Focus 2016-3b. Available online: https://ecommons.cornell.edu/bitstream/handle/1813/103660/Research-Focus-2016-3b-comparing-red-wine-color.pdf?sequence=1&isAllowed=y (accessed on 28 December 2021).
- Sun, L.; Fan, X.; Zhang, Y.; Jiang, J.; Sun, H.; Liu, C. Transcriptome analysis of genes involved in anthocyanins biosynthesis and transport in berries of black and white spine grapes (Vitis davidii). Hereditas 2016, 153, 1–22. [Google Scholar] [CrossRef] [Green Version]
- Dalbó, M.; Ye, G.; Weeden, N.; Steinkellner, H.; Sefc, K.; Reisch, B. A gene controlling sex in grapevines placed on a molecu-lar marker-based genetic map. Genome 2000, 43, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Riaz, S.; Dangl, G.; Edwards, K.; Meredith, C. A microsatellite marker based framework linkage map of Vitis vinifera L. Theor. Appl. Genet. 2004, 108, 864–872. [Google Scholar] [CrossRef] [PubMed]
- Lowe, K.; Walker, M. Genetic linkage map of the interspecific grape rootstock cross Ramsey (Vitis champinii)× Riparia Gloire (Vitis riparia). Theor. Appl. Genet. 2006, 112, 1582–1592. [Google Scholar] [CrossRef]
- Marguerit, E.; Boury, C.; Manicki, A.; Donnart, M.; Butterlin, G.; Némorin, A.; Wiedemann-Merdinoglu, S.; Merdinoglu, D.; Ollat, N.; Decroocq, S. Genetic dissection of sex determinism, inflorescence morphology and downy mildew resistance in grapevine. Theor. Appl. Genet. 2009, 118, 1261–1278. [Google Scholar] [CrossRef] [PubMed]
- Battilana, J.; Lorenzi, S.; Moreira, F.M.; Moreno-Sanz, P.; Failla, O.; Emanuelli, F.; Grando, M.S. Linkage mapping and molec-ular diversity at the flower sex locus in wild and cultivated grapevine reveal a prominent SSR haplotype in hermaphrodite plants. Mol. Biotechnol. 2013, 54, 1031–1037. [Google Scholar] [CrossRef] [Green Version]
- Picq, S.; Santoni, S.; Lacombe, T.; Latreille, M.; Weber, A.; Ardisson, M.; Ivorra, S.; Maghradze, D.; Arroyo-Garcia, R.; Chatelet, P. A small XY chromosomal region explains sex determination in wild dioecious V. vinifera and the reversal to hermaph-roditism in domesticated grapevines. BMC Plant Biol. 2014, 14, 229. [Google Scholar] [CrossRef] [Green Version]
- Massonnet, M.; Cochetel, N.; Minio, A.; Vondras, A.M.; Lin, J.; Muyle, A.; Garcia, J.F.; Zhou, Y.; Delledonne, M.; Riaz, S. The genetic basis of sex determination in grapes. Nat. Commun. 2020, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Lewter, J.; Worthington, M.L.; Clark, J.R.; Varanasi, A.V.; Nelson, L.; Owens, C.L.; Conner, P.; Gunawan, G. High-density linkage maps and loci for berry color and flower sex in muscadine grape (Vitis rotundifolia). Theor. Appl. Genet. 2019, 132, 1571–1585. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wang, N.; Fang, L.-C.; Liang, Z.-C.; Li, S.-H.; Wu, B.-H. Construction of a high-density genetic map and QTLs mapping for sugars and acids in grape berries. BMC Plant Biol. 2015, 15, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bayo-Canha, A.; Costantini, L.; Fernández-Fernández, J.I.; Martínez-Cutillas, A.; Ruiz-García, L. QTLs related to berry acidi-ty identified in a wine grapevine population grown in warm weather. Plant Mol. Biol. Rep. 2019, 37, 157–169. [Google Scholar] [CrossRef]
- Duchêne, É.; Dumas, V.; Butterlin, G.; Jaegli, N.; Rustenholz, C.; Chauveau, A.; Bérard, A.; le Paslier, M.C.; Gaillard, I.; Merdinoglu, D. Genetic variations of acidity in grape berries are controlled by the interplay between organic acids and potas-sium. Theor. Appl. Genet. 2020, 133, 993–1008. [Google Scholar] [CrossRef] [PubMed]
- Reshef, N.; Karn, A.; Manns, D.C.; Mansfield, A.K.; Cadle-Davidson, L.; Reisch, B.; Sacks, G.L. Stable QTL for malate levels in ripe fruit and their transferability across Vitis species. Hort. Res. 2022, 9, uhac009. [Google Scholar]
- Negus, K.L.; Chen, L.-L.; Fresnedo-Ramírez, J.; Scott, H.A.; Sacks, G.L.; Cadle-Davidson, L.; Hwang, C.-F. Identification of QTLs for berry acid and tannin in a Vitis aestivalis-derived‘Norton’-based population. Fruit Res. 2021, 1, 1–11. [Google Scholar] [CrossRef]
- Lin, J.; Massonnet, M.; Cantu, D. The genetic basis of grape and wine aroma. Hortic. Res. 2019, 6, 1–24. [Google Scholar] [CrossRef] [Green Version]
- Kalua, C.M.; Boss, P.K. Evolution of volatile compounds during the development of Cabernet Sauvignon grapes (Vitis vinifera L.). J. Agric. Food Chem. 2009, 57, 3818–3830. [Google Scholar] [CrossRef]
- Sun, Q.; Gates, M.J.; Lavin, E.H.; Acree, T.E.; Sacks, G.L. Comparison of odor-active compounds in grapes and wines from Vitis vinifera and non-foxy American grape species. J. Agric. Food Chem. 2011, 59, 10657–10664. [Google Scholar] [CrossRef]
- Ferrandino, A.; Carlomagno, A.; Baldassarre, S.; Schubert, A. Varietal and pre-fermentative volatiles during ripening of Vitis vinifera cv Nebbiolo berries from three growing areas. Food Chem. 2012, 135, 2340–2349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mozzon, M.; Savini, S.; Boselli, E.; Thorngate, J.H. The herbaceous character of wines. Ital. J. Food Sci. 2016, 28, 190–207. [Google Scholar] [CrossRef]
- Lei, Y.; Xie, S.; Guan, X.; Song, C.; Zhang, Z.; Meng, J. Methoxypyrazines biosynthesis and metabolism in grape: A review. Food Chem. 2018, 245, 1141–1147. [Google Scholar] [CrossRef] [PubMed]
- Guillaumie, S.; Ilg, A.; Réty, S.; Brette, M.; Trossat-Magnin, C.; Decroocq, S.; Léon, C.; Keime, C.; Ye, T.; Bal-tenweck-Guyot, R. Genetic analysis of the biosynthesis of 2-methoxy-3-isobutylpyrazine, a major grape-derived aroma compound impacting wine quality. Plant Physiol. 2013, 162, 604–615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dunlevy, J.D.; Dennis, E.G.; Soole, K.L.; Perkins, M.V.; Davies, C.; Boss, P.K. A methyltransferase essential for the methoxy-pyrazine-derived flavour of wine. Plant J. 2013, 75, 606–617. [Google Scholar] [CrossRef] [PubMed]
- Zuo, J.-F.; Niu, Y.; Cheng, P.; Feng, J.-Y.; Han, S.-F.; Zhang, Y.-H.; Shu, G.; Wang, Y.; Zhang, Y.-M. Effect of marker segrega-tion distortion on high density linkage map construction and QTL mapping in Soybean (Glycine max L.). Heredity 2019, 123, 579–592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coulton, A.; Przewieslik-Allen, A.M.; Burridge, A.J.; Shaw, D.S.; Edwards, K.J.; Barker, G.L. Segregation dis-tortion: Utilizing simulated genotyping data to evaluate statistical methods. PLoS ONE 2020, 15, e0228951. [Google Scholar] [CrossRef] [Green Version]
- Zhou, W.; Tang, Z.; Hou, J.; Hu, N.; Yin, T. Genetic map construction and detection of genetic loci underlying segregation distortion in an intraspecific cross of Populus deltoides. PLoS ONE 2015, 10, e0126077. [Google Scholar] [CrossRef] [Green Version]
- Garris, A.; Clark, L.; Owens, C.; McKay, S.; Luby, J.; Mathiason, K.; Fennell, A. Mapping of photoperiod-induced growth cessation in the wild grape Vitis riparia. J. Am. Soc. Hortic. Sci. 2009, 134, 261–272. [Google Scholar] [CrossRef]
- Yang, S.; Fresnedo-Ramírez, J.; Sun, Q.; Manns, D.C.; Sacks, G.L.; Mansfield, A.K.; Luby, J.J.; Londo, J.P.; Reisch, B.I.; Cadle-Davidson, L.E. Next generation mapping of enological traits in an F2 interspecific grapevine hybrid family. PLoS ONE 2016, 11, e0149560. [Google Scholar] [CrossRef] [Green Version]
- Lijavetzky, D.; Ruiz-García, L.; Cabezas, J.A.; de Andrés, M.T.; Bravo, G.; Ibáñez, A.; Carreño, J.; Cabello, F.; Ibáñez, J.; Mar-tínez-Zapater, J.M. Molecular genetics of berry colour variation in table grape. Mol. Genet. Genom. 2006, 276, 427–435. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, V.; Matus, J.T.; Pinto-Carnide, O.; Carrasco, D.; Arroyo-García, R.; Castro, I. Genetic analysis of a white-to-red berry skin color reversion and its transcriptomic and metabolic consequences in grapevine (Vitis vinifera cv.‘Moscatel Galego’). BMC Genom. 2019, 20, 952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coito, J.L.; Silva, H.; Ramos, M.J.; Montez, M.; Cunha, J.; Amâncio, S.; Costa, M.M.; Rocheta, M. Vitis flower sex specification acts downstream and independently of the ABCDE model genes. Front. Plant Sci. 2018, 9, 1029. [Google Scholar] [CrossRef] [PubMed]
- Zou, C.; Massonnet, M.; Minio, A.; Patel, S.; Llaca, V.; Karn, A.; Gouker, F.; Cadle-Davidson, L.; Reisch, B.; Fennell, A. Multi-ple independent recombinations led to hermaphroditism in grapevine. Proc. Natl. Acad. Sci. USA 2021, 118, 15. [Google Scholar] [CrossRef] [PubMed]
- Roman, T.; Barp, L.; Malacarne, M.; Nardin, T.; Nicolini, G.; Larcher, R. Mono-and di-glucoside anthocyanins extraction dur-ing the skin contact fermentation in hybrid grape varieties. Eur. Food Res. Technol. 2019, 245, 2373–2383. [Google Scholar] [CrossRef]
- OIV. International Code of Oenological Practices; International Code of Oenological Practices: Paris, France, 2015; ISBN 978-2-85038-030-3. [Google Scholar]
- Costantini, L.; Malacarne, G.; Lorenzi, S.; Troggio, M.; Mattivi, F.; Moser, C.; Grando, M.S. New candidate genes for the fine regulation of the colour of grapes. J. Exp. Bot. 2015, 66, 4427–4440. [Google Scholar] [CrossRef]
- Janvary, L.; Hoffmann, T.; Pfeiffer, J.; Hausmann, L.; Töpfer, R.; Fischer, T.C.; Schwab, W. A double mutation in the antho-cyanin 5-O-glucosyltransferase gene disrupts enzymatic activity in Vitis vinifera L. J. Agric. Food Chem. 2009, 57, 3512–3518. [Google Scholar] [CrossRef]
- He, F.; Chen, W.-K.; Yu, K.-J.; Ji, X.-N.; Duan, C.-Q.; Reeves, M.J.; Wang, J. Molecular and biochemical charac-terization of the UDP-glucose: Anthocyanin 5-O-glucosyltransferase from Vitis amurensis. Phytochemistry 2015, 117, 363–372. [Google Scholar] [CrossRef]
- Burzynski-Chang, E.A.; Brown, E.J.; Reshef, N.; Sacks, G.L. Malate content in wild Vitis spp. Demonstrates a range of behav-iors during berry maturation. Am. J. Enol. Vitic 2020, 71, 80–87. [Google Scholar] [CrossRef]
- Sweetman, C.; Deluc, L.G.; Cramer, G.R.; Ford, C.M.; Soole, K.L. Regulation of malate metabolism in grape berry and other developing fruits. Phytochemistry 2009, 70, 1329–1344. [Google Scholar] [CrossRef]
- De Angeli, A.; Baetz, U.; Francisco, R.; Zhang, J.; Chaves, M.M.; Regalado, A. The vacuolar channel VvALMT9 mediates malate and tartrate accumulation in berries of Vitis vinifera. Planta 2013, 238, 283–291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rienth, M.; Torregrosa, L.; Sarah, G.; Ardisson, M.; Brillouet, J.-M.; Romieu, C. Temperature desynchronizes sugar and organ-ic acid metabolism in ripening grapevine fruits and remodels their transcriptome. BMC Plant Biol. 2016, 16, 164. [Google Scholar] [CrossRef] [PubMed]
- Fanizza, G.; Lamaj, F.; Costantini, L.; Chaabane, R.; Grando, M. QTL analysis for fruit yield components in table grapes (Vitis vinifera). Theor. Appl. Genet. 2005, 111, 658–664. [Google Scholar] [CrossRef] [PubMed]
- Costantini, L.; Battilana, J.; Lamaj, F.; Fanizza, G.; Grando, M.S. Berry and phenology-related traits in grapevine (Vitis vinifera L.): From quantitative trait loci to underlying genes. BMC Plant Biol. 2008, 8, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grzeskowiak, L.; Costantini, L.; Lorenzi, S.; Grando, M.S. Candidate loci for phenology and fruitfulness con-tributing to the phenotypic variability observed in grapevine. Theor. Appl. Genet. 2013, 126, 2763–2776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lacey, M.J.; Allen, M.S.; Harris, R.L.; Brown, W.V. Methoxypyrazines in Sauvignon blanc grapes and wines. Am. J. Enol. Vitic. 1991, 42, 103–108. [Google Scholar]
- Slegers, A.; Angers, P.; Pedneault, K. Volatile compounds from must and wines from five white grape varieties. J. Food Chem. Nanotechnol. 2017, 3, 8–17. [Google Scholar] [CrossRef]
- Rahman, F.U.; Nawaz, M.A.; Liu, R.; Sun, L.; Jiang, J.; Fan, X.; Liu, C.; Zhang, Y. Evaluation of volatile aroma compounds from Chinese wild grape berries by headspace-SPME with GC-MS. Food Sci. Technol. 2022, 42, e54320. [Google Scholar] [CrossRef]
- Pilati, S.; Brazzale, D.; Guella, G.; Milli, A.; Ruberti, C.; Biasioli, F.; Zottini, M.; Moser, C. The onset of grapevine berry ripen-ing is characterized by ROS accumulation and lipoxygenase-mediated membrane peroxidation in the skin. BMC Plant Biol. 2014, 14, 87. [Google Scholar] [CrossRef] [Green Version]
- Pilati, S.; Perazzolli, M.; Malossini, A.; Cestaro, A.; Demattè, L.; Fontana, P.; dal Ri, A.; Viola, R.; Velasco, R.; Moser, C. Genome-wide transcriptional analysis of grapevine berry ripening reveals a set of genes similarly modulated during three seasons and the occurrence of an oxidative burst at veraison. BMC Genom. 2007, 8, 428. [Google Scholar] [CrossRef] [Green Version]
- Fennell, A.; Mathiason, K.; Luby, J. Genetic Segregation for Indicators of Photoperiod Control of Dormancy In-Duction in Vitis Species. In Proceedings of the VII International Symposium on Grapevine Physiology and Biotechnology, Davis, CA, USA, 31 August 2005; pp. 533–540. [Google Scholar]
- Van Ooijen, J. Multipoint maximum likelihood mapping in a full-sib family of an outbreeding species. Genet. Res. 2011, 93, 343–349. [Google Scholar] [CrossRef] [PubMed]
- Voorrips, R.R. MapChart: Software for the Graphical Presentation of Linkage Maps and QTLs. J. Hered. 2002, 93, 77r78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mesonet. Real Weather Data from the Ground up. Available online: Sdstate.edu/mesonet (accessed on 1 November 2021).
- Manns, D.C.; Mansfield, A.K. A core–shell column approach to a comprehensive high-performance liquid chromatography phenolic analysis of Vitis vinifera L. and interspecific hybrid grape juices, wines, and other matrices following either solid phase extraction or direct injection. J. Chromatogr. A 2012, 1251, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Burzynski-Chang, E.A.; Ryona, I.; Reisch, B.I.; Gonda, I.; Foolad, M.R.; Giovannoni, J.J.; Sacks, G.L. HS-SPME-GC-MS anal-yses of volatiles in plant populations—Quantitating compound× individual matrix effects. Molecules 2018, 23, 2436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Revelle, W. Psych: Procedures for Psychological, Psychometric, and Personality Research; R Package Version 2.1.9; Northwestern University: Evanston, IL, USA, 2018; Available online: https://CRAN.R-project.org/package=psych (accessed on 28 December 2021).
- Team, R.C. A language and Environment for Statistical Computing; R. Foundation: Vienna, Austria, 2021. [Google Scholar]
- Broman, K.W.; Wu, H.; Sen, Ś.; Churchill, G.A. R/qtl: QTL mapping in experimental crosses. Bioinformatics 2003, 19, 889–890. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venables, W.; Ripley, B. Modern Applied Statistics with S, 4th ed.; Springer: New York, NY, USA, 2002. [Google Scholar]
- Peña, E.A.; Slate, E.H. Global validation of linear model assumptions. J. Am. Stat. Assoc. 2006, 101, 341–354. [Google Scholar] [CrossRef]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.; Potter, S.C.; Finn, R.D. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef] [Green Version]






| Parameter | Value |
|---|---|
| Number of F2 genotypes | 514 |
| Total (GBS and rhAmpSeq) markers used in map curation | 3428 |
| Markers in LGs | 2519 |
| GBS markers in LGs | 1449 |
| rhAmpSeq markers in LGs | 1070 |
| LGs | 19 |
| Distortion threshold | 1 × 10−21 |
| Number of markers that formed different LG | 0 |
| Mismapped markers in LG | 0 |
| Markers not in any LG | 0 |
| Problematic markers in LG | 0 |
| Non-informative marker | 677 |
| Distorted marker numbers | 232 |
| Genetic map size (cM) | 1131.3 |
| Genome-wide recombination rate (cM/Mb) | 2.5 |
| Average distance between markers (cM) | 0.5 |
| Genome coverage (%) | 96.3 |
| Largest gap (cM) | 11.3 |
| Chromosome | Number of Markers | Chromosome Genetic Length (cM) | Average Spacing (cM) | Maximum spacing (cM) | Correlation (Spearman) |
|---|---|---|---|---|---|
| 1 | 150 | 65.3 | 0.4 | 4.7 | 0.9999 * |
| 2 | 112 | 52.2 | 0.5 | 4.2 | 0.9998 * |
| 3 | 97 | 48.2 | 0.5 | 5.3 | 0.9996 * |
| 4 | 181 | 62.7 | 0.3 | 2.3 | 0.9999 * |
| 5 | 193 | 62.5 | 0.3 | 2.0 | 0.9999 * |
| 6 | 131 | 60.1 | 0.5 | 4.9 | 0.9998 * |
| 7 | 176 | 79.1 | 0.5 | 8.1 | 0.9999 * |
| 8 | 145 | 59.3 | 0.4 | 2.8 | 0.9994 * |
| 9 | 117 | 60.0 | 0.5 | 4.2 | 0.9999 * |
| 10 | 88 | 55.9 | 0.6 | 11.3 | 0.9997 * |
| 11 | 105 | 63.9 | 0.6 | 4.4 | 0.9996 * |
| 12 | 124 | 56.5 | 0.5 | 3.3 | 0.9999 * |
| 13 | 153 | 68.7 | 0.5 | 4.9 | 0.9999 * |
| 14 | 161 | 68.9 | 0.4 | 3.1 | 0.9999 * |
| 15 | 34 | 18.9 | 0.6 | 2.1 | 0.9997 * |
| 16 | 114 | 55.5 | 0.5 | 3.1 | 0.9994 * |
| 17 | 140 | 59.4 | 0.4 | 4.6 | 0.9998 * |
| 18 | 192 | 79.9 | 0.4 | 8.5 | 0.9999 * |
| 19 | 106 | 54.3 | 0.5 | 4.8 | 0.9999 * |
| Overall | 2519 | 1131.3 | 0.5 |
| Trait | Chromosome | LOD | Peak Position (Physical Position (Mb)) | R2 | Physical (Mb) Position at 95% Bayesian Interval |
|---|---|---|---|---|---|
| Berry color | 2 | 22.8 | 14.11 | NP | 11.00:17.48 |
| Flower type | 2 | 17.2 | 4.65 | NP | 4.50:4.70 |
| Total anthocyanin 2013 | 2 | 9.4 | 13.54 | NP | 6.97:17.85 |
| Total anthocyanin 2018 | 2 | 10.2 | 13.54 | NP | 6.97:17.85 |
| Total anthocyanin 2013 | 18 | 3.4 | 6.94 | NP | 6.23:10.21 |
| Total monoglucosides 2013 | 2 | 12.2 | 9.13 | NP | 8.58:14.87 |
| Total monoglucosides 2018 | 2 | 3.8 | 5.87 | NP | 2.99:16:74 |
| Total diglucosides 2013 | 2 | 5.7 | 8.11 | NP | 7.41:9.07 |
| Total diglucosides 2013 | 9 | 4.8 | 6.19 | NP | 0.89:6.99 |
| Total diglucosides 2018 | 9 | 3.2 | 6.52 | NP | 3.74:9.57 |
| Malic acid 2013 | 6 | 4.5 | 11.92 | 28.2 | 2.31:15.48 |
| Malic acid 2016 | 6 | 4.1 | 7.86 | 23.8 | 2.55:18.52 |
| Malic acid 2018 a | 6 | 3.8 | 5.59 | 22.9 | 0.91:8.28 |
| Malic acid 2018 | 1 | 5.2 | 20.62 | 31.0 | 18.88:23.67 |
| Titratable acidity 2016 a | 1 | 3.8 | 6.29 | 22.5 | 0.97:8.38 |
| Titratable acidity 2018 | 1 | 4.5 | 19.66 | 29.1 | 7.24:23.7 |
| Titratable acidity 2013 | 6 | 4.8 | 15.25 | 29.7 | 2.85:16.63 |
| Titratable acidity 2016 | 6 | 4.2 | 7.86 | 24.4 | 2.76:18.14 |
| Titratable acidity 2018 | 6 | 4.1 | 5.59 | 28.1 | 0.28:18.52 |
| (E)-2-hexenal 2013 | 2 | 4.4 | 7.47 | 29.4 | 5.35:18.70 |
| (E)-2-hexenal 2018 a | 2 | 4.0 | 4.83 | 27.8 | 0.18:17.48 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alahakoon, D.; Fennell, A.; Helget, Z.; Bates, T.; Karn, A.; Manns, D.; Mansfield, A.K.; Reisch, B.I.; Sacks, G.; Sun, Q.; et al. Berry Anthocyanin, Acid, and Volatile Trait Analyses in a Grapevine-Interspecific F2 Population Using an Integrated GBS and rhAmpSeq Genetic Map. Plants 2022, 11, 696. https://doi.org/10.3390/plants11050696
Alahakoon D, Fennell A, Helget Z, Bates T, Karn A, Manns D, Mansfield AK, Reisch BI, Sacks G, Sun Q, et al. Berry Anthocyanin, Acid, and Volatile Trait Analyses in a Grapevine-Interspecific F2 Population Using an Integrated GBS and rhAmpSeq Genetic Map. Plants. 2022; 11(5):696. https://doi.org/10.3390/plants11050696
Chicago/Turabian StyleAlahakoon, Dilmini, Anne Fennell, Zachary Helget, Terry Bates, Avinash Karn, David Manns, Anna Katharine Mansfield, Bruce I. Reisch, Gavin Sacks, Qi Sun, and et al. 2022. "Berry Anthocyanin, Acid, and Volatile Trait Analyses in a Grapevine-Interspecific F2 Population Using an Integrated GBS and rhAmpSeq Genetic Map" Plants 11, no. 5: 696. https://doi.org/10.3390/plants11050696