In Vitro Evaluation of the Inhibitory Activity of Different Selenium Chemical Forms on the Growth of a Fusarium proliferatum Strain Isolated from Rice Seedlings
Abstract
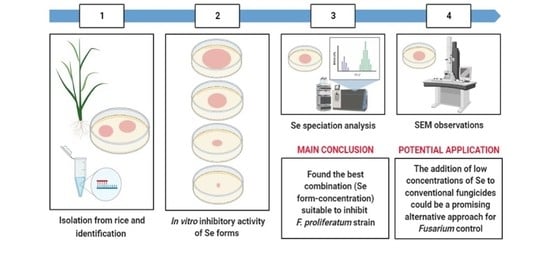
:1. Introduction
2. Results
2.1. Identification of the F. proliferatum Strain PG-CH1 Isolated from Rice Seedlings
2.2. The In Vitro Inhibitory Activity of Different Se Forms on F. proliferatum Strain PG-CH1 Growth
2.3. Se Speciation
2.4. Selenium (20 mg kg−1) from Sodium Selenite Effect Observed by SEM
3. Discussion
- (i)
- The occurrence of redox processes mediated by Se (e.g., selenate reduction to selenite, see Figure 4b), which may have affected the regular physiological processes of the fungal microorganism.
- (ii)
- The formation of organic Se compounds caused by the fungus (e.g., inorganic Se is converted to Se-Met and Se-Cys, see Figure 4a,b; Se-Met is converted to Se-Cys, see Figure 4c; Se-Cys is converted to Se-Met in small amounts, see Figure 4d), which may lead to direct incorporation of Se-Cys and Se-Met into proteins, rather than the analog sulfur-containing amino acids S-Cys and S-Met, a process previously reported in the literature [53,54]. In this case, the growth inhibitory effect may be ascribed to a different conformation of the Se proteins concerning analog S proteins to such an extent that a modification in protein activity should not be excluded.
- (iii)
- The direct incorporation of the applied Se-Cys and Se-Met into proteins, a process which may explain the higher bioavailability of organic Se forms compared to inorganic Se forms.
- (iv)
- The relationship of Se with oxidative stress [5]. For example, it is known that in certain filamentous fungi, mycelium metamorphosis, in structures that ensure fungal propagation, is induced by increased oxidative stress. Conversely, decreased oxidative stress causes a permanence of undifferentiated mycelia, and inhibition of metamorphosis and fungal propagation [55]. The extension of this theory predicts that any antioxidant (such as Se at certain doses) can stop fungal propagation by inhibiting its metamorphosis, thus acting as a natural fungicide.
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Obtainment of Fusarium proliferatum Strain PG-CH1 from Rice Seedlings
4.3. In Vitro Evaluation of the Inhibitory Activity of Se Forms on F. proliferatum Strain PG-CH1
(radial growthcontrol − radial growthtreatment): radial growthcontrol × 100
4.4. Se Speciation Analysis
4.5. Observation by Scanning Electron Microscopy
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Steinbrenner, H.; Speckmann, B.; Klotz, L.O. Selenoproteins: Antioxidant selenoenzymes and beyond. Arch. Biochem. Biophys. 2016, 595, 113–119. [Google Scholar] [CrossRef]
- Roman, M.; Jitaru, P.; Barbante, C. Selenium biochemistry and its role for human health. Metallomics 2014, 6, 25–54. [Google Scholar] [CrossRef]
- D’Amato, R.; Regni, L.; Falcinelli, B.; Mattioli, S.; Benincasa, P.; Dal Bosco, A.; Pacheco, P.; Proietti, P.; Troni, E.; Santi, C.; et al. Current knowledge on selenium biofortification to improve the nutraceutical profile of food: A comprehensive review. J. Agric. Food Chem. 2020, 68, 4075–4097. [Google Scholar] [CrossRef]
- Ursini, F.; Maiorino, M. Glutathione peroxidases. In Encyclopedia of Biological Chemistry, 2nd ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2013; pp. 399–404. [Google Scholar]
- Gupta, M.; Gupta, S. An overview of selenium uptake, metabolism, and toxicity in plants. Front Plant. Sci. 2017, 7, 2074. [Google Scholar] [CrossRef] [Green Version]
- Vats, S. Biotic and Abiotic Stress Tolerance in Plants; Vats, S., Ed.; Springer: Singapore; Springer Nature Singapore Pte Ltd.: Singapore, 2018. [Google Scholar]
- Hasanuzzaman, M.; Bhuyan, M.H.M.B.; Raza, A.; Hawrylak-Nowak, B.; Matraszek-Gawron, R.; Mahmud, J.A.; Nahar, K.; Fujita, M. Selenium in plants: Boon or bane? Environ. Exp. Bot. 2020, 178, 104170. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Borhannuddin Bhuyan, M.H.M.; Raza, A.; Hawrylak-Nowak, B.; Matraszek-Gawron, R.; Nahar, K.; Fujita, M. Selenium toxicity in plants and environment: Biogeochemistry and remediation possibilities. Plants 2020, 9, 1711. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsov, V.; Kuznetsov, V. Selenium regulates the water status of plants exposed to drought. Dokl. Biol. Sci. 2003, 390, 266–268. [Google Scholar] [CrossRef] [PubMed]
- Gilmara, P.d.S.; Leonardo, C.C.; Victor, V.C.; Sylvia, L.O.S.; Edilaine, I.F.T. Selenium and agricultural crops. Afr. J. Agric. Res. 2017, 12, 2545–2554. [Google Scholar] [CrossRef] [Green Version]
- Bocchini, M.; D’Amato, R.; Ciancaleoni, S.; Fontanella, M.C.; Palmerini, C.A.; Beone, G.M.; Onofri, A.; Negri, V.; Marconi, G.; Albertini, E.; et al. Soil Selenium (Se) biofortification changes the physiological, biochemical and epigenetic responses to water stress in Zea mays L. by inducing a higher drought tolerance. Front. Plant Sci. 2018, 9, 389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Proietti, P.; Nasini, L.; Del Buono, D.; D’Amato, R.; Tedeschini, E.; Businelli, D. Selenium protects olive (Olea europaea L.) from drought stress. Sci. Hortic. 2013, 164, 165–171. [Google Scholar] [CrossRef]
- Feng, R.; Wei, C.; Tu, S. The roles of selenium in protecting plants against abiotic stresses. Environ. Exp. Bot. 2013, 87, 58–68. [Google Scholar] [CrossRef]
- Rayman, M.P. Selenium intake, status, and health: A complex relationship. Hormones 2019, 19, 9–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thiry, C.; Ruttens, A.; De Temmerman, L.; Schneider, Y.J.; Pussemier, L. Current knowledge in species-related bioavailability of selenium in food. Food Chem. 2012, 130, 767–784. [Google Scholar] [CrossRef]
- Bhatia, P.; Aureli, F.; D’Amato, M.; Prakash, R.; Cameotra, S.S.; Nagaraja, T.P.; Cubadda, F. Selenium bioaccessibility and speciation in biofortified Pleurotus mushrooms grown on selenium-rich agricultural residues. Food Chem. 2013, 140, 225–230. [Google Scholar] [CrossRef]
- Hasanuzzam, M.; Hossain, M.A.; Fujita, M. Selenium in higher plants: Physiological role, antioxidant metabolism and abiotic stress tolerance. J. Plant Sci. 2010, 5, 354–375. [Google Scholar] [CrossRef] [Green Version]
- Wu, Z.-L.; Yin, X.-B.; Lin, Z.-Q.; Bañuelos, G.S.; Yuan, L.-X.; Liu, Y.; Li, M. Inhibitory effect of selenium against Penicillium expansum and its possible mechanisms of action. Curr. Microbiol. 2014, 69, 192–201. [Google Scholar] [CrossRef] [PubMed]
- Razak, A.A.; El-Tantawy, H.; El-Sheikh, H.H.; Gharieb, M.M. Influence of selenium on the efficiency of fungicide action against certain fungi. Biol. Trace Elem. Res. 1991, 28, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Ramadan, S.E.; Razak, A.A.; Yousseff, Y.A.; Sedky, N.M. Selenium metabolism in a strain of Fusarium. Biol. Trace Elem. Res. 1988, 18, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Hua, C.; Yang, L.; Zhang, Y.; Sun, Z.; Li, L.; Li, T. The effects of selenium on wheat fusarium head blight and DON accumulation were selenium compound-dependent. Toxins 2020, 12, 573. [Google Scholar] [CrossRef] [PubMed]
- Hanson, B.; Garifullina, G.F.; Lindblom, S.D.; Wangeline, A.; Ackley, A.; Kramer, K.; Norton, A.P.; Lawrence, C.B.; Pilon-Smits, E.A.H. Selenium accumulation protects Brassica juncea from invertebrate herbivory and fungal infection. New Phytol. 2003, 159, 461–469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Companioni, B.; Medrano, J.; Torres, J.A.; Flores, A.; Rodríguez, E.; Benavides, A. Protective action of sodium selenite against Fusarium wilt in tomato: Total protein contents, levels of phenolic compounds and changes in antioxidant potential. Acta Hortic. 2012, 947, 321–327. [Google Scholar] [CrossRef]
- You, J.; Zhang, J.; Wu, M.; Yang, L.; Chen, W.; Li, G. Multiple criteria-based screening of Trichoderma isolates for biological control of Botrytis cinerea on tomato. Biol. Control. 2016, 101, 31–38. [Google Scholar] [CrossRef]
- Filek, M.; Łabanowska, M.; Kurdziel, M.; Sieprawska, A. Electron paramagnetic resonance (EPR) spectroscopy in studies of the protective effects of 24-epibrasinoide and selenium against zearalenone-stimulation of the oxidative stress in germinating grains of wheat. Toxins 2017, 9, 178. [Google Scholar] [CrossRef] [PubMed]
- Kornaś, A.; Filek, M.; Sieprawska, A.; Bednarska-Kozakiewicz, E.; Gawrońska, K.; Miszalski, Z. Foliar application of selenium for protection against the first stages of mycotoxin infection of crop plant leaves. J. Sci. Food Agric. 2019, 99, 482–485. [Google Scholar] [CrossRef] [PubMed]
- Agar, G.; Alpsoy, L.; Bozari, S.; Erturk, F.A.; Yildirim, N. Determination of protective role of selenium against aflatoxin B1-induced DNA damage. Toxicol. Ind. Health 2013, 29, 396–403. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhao, Z.; Duan, B.; Hu, C.; Zhao, X.; Guo, Z. Effect of applied sulphur on the uptake by wheat of selenium applied as selenite. Plant Soil 2015, 386, 35–45. [Google Scholar] [CrossRef]
- D’Amato, R.; Fontanella, M.C.; Falcinelli, B.; Beone, G.M.; Bravi, E.; Marconi, O.; Benincasa, P.; Businelli, D. Selenium biofortification in rice (Oryza sativa L.) sprouting: Effects on Se yield and nutritional traits with focus on phenolic acid profile. J. Agric. Food Chem. 2018, 66, 4082–4090. [Google Scholar] [CrossRef]
- Kane, C.D.; Jasoni, R.L.; Peffley, E.P.; Thompson, L.D.; Green, C.J.; Pare, P.; Tissue, D. Nutrient solution and solution pH influences on onion growth and mineral content. J. Plant Nutr. 2006, 29, 375–390. [Google Scholar] [CrossRef]
- O’Donnell, K.; Nirenberg, H.I.; Aoki, T.; Cigelnik, E. A Multigene phylogeny of the Gibberella fujikuroi species complex: Detection of additional phylogenetically distinct species. Mycoscience 2000, 41, 61–78. [Google Scholar] [CrossRef]
- O’Donnell, K.; Rooney, A.P.; Proctor, R.H.; Brown, D.W.; McCormick, S.P.; Ward, T.J.; Frandsen, R.J.N.; Lysøe, E.; Rehner, S.A.; Aoki, T.; et al. Phylogenetic analyses of RPB1 and RPB2 support a middle Cretaceous origin for a clade comprising all agriculturally and medically important fusaria. Fungal Genet. Biol. 2013, 52, 20–31. [Google Scholar] [CrossRef]
- Palacios, S.A.; Susca, A.; Haidukowski, M.; Stea, G.; Cendoya, E.; Ramírez, M.L.; Chulze, S.N.; Farnochi, M.C.; Moretti, A.; Torres, A.M. Genetic variability and fumonisin production by Fusarium proliferatum isolated from durum wheat grains in Argentina. Int. J. Food Microbiol. 2015, 201, 35–41. [Google Scholar] [CrossRef]
- Chulze, S.N.; Ramirez, M.L.; Farnochi, M.C.; Pascale, M.; Visconti, A.; March, G. Fusarium and fumonisin occurrence in Argentinian corn at different ear maturity stages. J. Agric. Food Chem. 1996, 44, 2797–2801. [Google Scholar] [CrossRef]
- Amatulli, M.T.; Spadaro, D.; Gullino, M.L.; Garibaldi, A. Molecular identification of Fusarium spp. associated with bakanae disease of rice in Italy and assessment of their pathogenicity. Plant Pathol. 2010, 59, 839–844. [Google Scholar] [CrossRef]
- Wulff, E.G.; Sørensen, J.L.; Lübeck, M.; Nielsen, K.F.; Thrane, U.; Torp, J. Fusarium spp. associated with rice Bakanae: Ecology, genetic diversity, pathogenicity and toxigenicity. Environ. Microbiol. 2010, 12, 649–657. [Google Scholar] [CrossRef] [PubMed]
- Leslie, J.F.; Zeller, K.A.; Lamprecht, S.C.; Rheeder, J.P.; Marasas, W.F.O. Toxicity, pathogenicity, and genetic differentiation of five species of Fusarium from sorghum and millet. Phytopathology 2005, 95, 275–283. [Google Scholar] [CrossRef] [Green Version]
- Vismer, H.F.; Shephard, G.S.; van der Westhuizen, L.; Mngqawa, P.; Bushula-Njah, V.; Leslie, J.F. Mycotoxins produced by Fusarium proliferatum and F. pseudonygamai on maize, sorghum and pearl millet grains in vitro. Int. J. Food Microbiol. 2019, 296, 31–36. [Google Scholar] [CrossRef]
- Logrieco, A.; Doko, B.; Moretti, A.; Frisullo, S.; Visconti, A. Occurrence of fumonisin B1 and B2 in Fusarium proliferatum infected asparagus plants. J. Agric. Food Chem. 1998, 46, 5201–5204. [Google Scholar] [CrossRef]
- Gálvez, L.; Urbaniak, M.; Waśkiewicz, A.; Stępień, Ł.; Palmero, D. Fusarium proliferatum–Causal agent of garlic bulb rot in Spain: Genetic variability and mycotoxin production. Food Microbiol. 2017, 67, 41–48. [Google Scholar] [CrossRef]
- Abdalla, M.Y.; Al-Rokibah, A.; Moretti, A.; Mulè, G. Pathogenicity of toxigenic Fusarium proliferatum from date palm in Saudi Arabia. Plant Dis. 2000, 84, 321–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desjardins, A.E.; Manandhar, H.K.; Plattner, R.D.; Manandhar, G.G.; Poling, S.M.; Maragos, C.M. Fusarium species from nepalese rice and production of mycotoxins and gibberellic acid by selected species. Appl. Environ. Microbiol. 2000, 66, 1020–1025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohd Zainudin, N.A.I.; Razak, A.A.; Salleh, B. Bakanae disease of rice in malaysia and indonesia: Etiology of the causal agent based on morphological, physiological and pathogenicity characteristics. J. Plant Prot. Res. 2008, 48, 475–485. [Google Scholar] [CrossRef]
- Ji, F.; He, D.; Olaniran, A.O.; Mokoena, M.P.; Xu, J.; Shi, J. Occurrence, toxicity, production and detection of Fusarium mycotoxin: A review. Food Prod. Process. Nutr. 2019, 1, 1–14. [Google Scholar] [CrossRef]
- Braun, M.S.; Wink, M. Exposure, occurrence, and chemistry of fumonisins and their cryptic derivatives. Compr. Rev. Food Sci. Food Saf. 2018, 17, 769–791. [Google Scholar] [CrossRef] [Green Version]
- Schiavon, M.; Nardi, S.; dalla Vecchia, F.; Ertani, A. Selenium biofortification in the 21st century: Status and challenges for healthy human nutrition. Plant Soil 2020, 453, 245–270. [Google Scholar] [CrossRef] [PubMed]
- Reddy, C.C.; Massaro, E.J. Biochemistry of selenium: A brief overview. Toxicol. Sci. 1983, 3, 431–436. [Google Scholar] [CrossRef]
- McNeal, J.M.; Balistrieri, L.S. Geochemistry and Occurrence of Selenium: An Overview. In Selenium in Agriculture and the Environment; Jacob, L.W., Ed.; Soil Science Society of America: Madison, WI, USA, 1989; pp. 1–13. [Google Scholar]
- Boyd, R. Selenium stories. Nat. Chem. 2011, 3, 570. [Google Scholar] [CrossRef]
- Wu, Z.; Yin, X.; Bañuelos, G.S.; Lin, Z.Q.; Zhu, Z.; Liu, Y.; Yuan, L.; Li, M. Effect of selenium on control of postharvest gray mold of tomato fruit and the possible mechanisms involved. Front. Microbiol. 2016, 6, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Zhang, Y.; Zhang, F.; Hu, J.T.; Zhao, Y.M.; Cheng, B.L. Effects of selenium on Fusarium growth and associated fermentation products and the relationship with chondrocyte viability. Biomed. Environ. Sci. 2017, 30, 134–138. [Google Scholar] [PubMed]
- Jacob, C.; Giles, G.I.; Giles, G.I.; Sies, H. Sulfur and selenium: The role of oxidation state in protein structure and function. Angew. Chem. Int. Ed. 2003, 42, 4742–4758. [Google Scholar] [CrossRef] [PubMed]
- Läuchli, A. Selenium in plants: Uptake, functions, and environmental toxicity. Bot. Acta 1993, 106, 455–468. [Google Scholar] [CrossRef]
- Patching, S.G.; Gardiner, P.H.E. Recent developments in selenium metabolism and chemical speciation: A review. J. Trace Elem. Med. Biol. 1999, 13, 193–214. [Google Scholar] [CrossRef]
- Georgiou, C.D.; Patsoukis, N.; Papapostolou, I.; Zervoudakis, G. Sclerotial metamorphosis in filamentous fungi is induced by oxidative stress. Integr. Comp. Biol. 2006, 46, 691–712. [Google Scholar] [CrossRef]
- Brown, T.A.; Shrift, A. Selenium: Toxicity and tolerance in higher plants. Biol. Rev. 1982, 57, 59–84. [Google Scholar] [CrossRef]
- da Silva, M.d.C.S.; da Luz, J.M.R.; Paiva, A.P.S.; Mendes, D.R.; Carvalho, A.A.C.; Naozuka, J.; Kasuya, M.C.M. Growth and Tolerance of Pleurotus ostreatus at Different Selenium Forms. J. Agric. Sci. 2019, 11, 151. [Google Scholar] [CrossRef]
- Herrero, E.; Wellinger, R.E. Yeast as a model system to study metabolic impact of selenium compounds. Microb. Cell 2015, 2, 139–149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rao, Y.; McCooeye, M.; Windust, A.; Bramanti, E.; D’Ulivo, A.; Mester, Z. Mapping of selenium metabolic pathway in yeast by Liquid Chromatography−Orbitrap Mass Spectrometry. Anal. Chem. 2010, 82, 8121–8130. [Google Scholar] [CrossRef] [Green Version]
- Arnaudguilhem, C.; Bierla, K.; Ouerdane, L.; Preud’homme, H.; Yiannikouris, A.; Lobinski, R. Selenium metabolomics in yeast using complementary reversed-phase/hydrophilic ion interaction (HILIC) liquid chromatography-electrospray hybrid quadrupole trap/Orbitrap mass spectrometry. Anal. Chim. Acta 2012, 757, 26–38. [Google Scholar] [CrossRef] [PubMed]
- Rosenfield, C.E.; Kenyon, J.A.; James, B.R.; Santelli, C.M. Selenium (IV, VI) reduction and tolerance by fungi in an oxic environment. Geobiology 2016, 15, 441–452. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Liu, R.; Gong, X.; Li, Z.; Huang, Q.; Wang, H.; Song, G. Synthesis and herbicidal activity of N,N-diethyl-3-(arylselenonyl)-1 H-1,2,4-triazole-1 -carboxamide. J. Agric. Food Chem. 2006, 54, 7724–7728. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.K. Synthesis and fungicidal activity of novel 3-(substituted/unsubstituted phenylselenonyl)-1-ribosyl/deoxyribosyl-1 H -1,2,4-triazole. J. Agric. Food Chem. 2012, 60, 5813–5818. [Google Scholar] [CrossRef]
- Zhu, Y.G.; Pilon-Smits, E.A.H.; Zhao, F.J.; Williams, P.N.; Meharg, A.A. Selenium in higher plants: Understanding mechanisms for biofortification and phytoremediation. Trends Plant Sci. 2009, 14, 436–442. [Google Scholar] [CrossRef] [PubMed]
- Hartikainen, H. Biogeochemistry of selenium and its impact on food chain quality and human health. J. Trace Elem. Med. Biol. 2005, 18, 309–318. [Google Scholar] [CrossRef] [PubMed]
- Beccari, G.; Senatore, M.T.; Tini, F.; Sulyok, M.; Covarelli, L. Fungal community, Fusarium head blight complex and secondary metabolites associated with malting barley grains harvested in Umbria, central Italy. Int. J. Food Microbiol. 2018, 273, 33–42. [Google Scholar] [CrossRef]
- Beccari, G.; Prodi, A.; Senatore, M.T.; Balmas, V.; Tini, F.; Onofri, A.; Pedini, L.; Sulyok, M.; Brocca, L.; Covarelli, L. Cultivation area affects the presence of fungal communities and secondary metabolites in Italian durum wheat grains. Toxins 2020, 12, 97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Donnell, K.; Kistler, H.C.; Cigelnik, E.; Ploetz, R.C. Multiple evolutionary origins of the fungus causing panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. USA 1998, 95, 2044–2049. [Google Scholar] [CrossRef] [Green Version]
- National Center for Biotechnology Information (NCBI) Basic Local Search (BLAST) Tool. Available online: http://blast.ncbi.nlm.nih.gov (accessed on 1 November 2018).
- Amatulli, M.T.; Spadaro, D.; Gullino, M.L.; Garibaldi, A. Conventional and real-time PCR for the identification of Fusarium fujikuroi and Fusarium proliferatum from diseased rice tissues and seeds. Eur. J. Plant Pathol. 2012, 134, 401–408. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- O’Donnell, K.; McCormick, S.P.; Busman, M.; Proctor, R.H.; Ward, T.J.; Doehring, G.; Geiser, D.M.; Alberts, J.F.; Rheeder, J.P.; Marasas; et al. 1984 “Toxigenic Fusarium species: Identity and mycotoxicology” revisited. Mycologia 2018, 110, 1058–1080. [Google Scholar] [CrossRef]
- Alves, E.; Lucas, G.C.; Pozza, E.A.; de Carvalho Alves, M. Scanning electron microscopy for fungal sample examination. In Laboratory Protocols in Fungal Biology; Springer: New York, NY, USA, 2013; pp. 133–150. [Google Scholar]
- Onofri, A.; Pannacci, E. Spreadsheet tools for biometry classes in crop science programmes. Commun. Biometry Crop. Sci. 2014, 9, 3–13. [Google Scholar]





| Strain | Species | Species Complex | Host | Geographic Origin/Substrate | GenBank Accession Number | References |
|---|---|---|---|---|---|---|
| MRC 548 | F. verticillioides | Fujikuroi | Maize | South Africa | MH582323.1 | [72] |
| MRC 602 | F. verticillioides | Fujikuroi | Maize | South Africa | MH582331.1 | [72] |
| MRC 1411 | F. tapsinum | Fujikuroi | Maize | North Carolina-USA | MH582337.1 | [72] |
| MRC 1784 | F. fujikuroi | Fujikuroi | Rawcotton | Georgia, USA | MH582338.1 | [72] |
| MRC 2324 | F. proliferaum | Fujikuroi | Cotton boll | Alabama, USA | MH582344.1 | [72] |
| MRC 2387 | F. fujikuroi | Fujikuroi | Rice | Japan | MH582340.1 | [70] |
| MRC 2390 | F. fujikuroi | Fujikuroi | Unknown | Unknown | MH582342.1 | [72] |
| MRC 2535 | F. proliferatum | Fujikuroi | River sediment | Japan | MH582346.1 | [72] |
| MRC 2629 | F. verticillioides | Fujikuroi | Maize | Iowa, USA | MH582328.1 | [72] |
| MRC 2633 | F. proliferatum | Fujikuroi | Wheat | India | MH582345.1 | [72] |
| S1S | F. proliferatum | Fujikuroi | Rice (seed) | Italy | JN092349 | [70] |
| 2–27 | F. proliferatum | Fujikuroi | Rice (seed) | Italy | JN092351 | [70] |
| M3096 | F. fujikuroi | Fujikuroi | Rice | Georgia, USA | JN092356 | [70] |
| M1150 | F. fujikuroi | Fujikuroi | Rice | Taiwan | JN092354 | [70] |
| G18SXS9-2 | F. proliferatum | Fujikuroi | Unknown | Unknown | MK952837.1 | Unknown |
| MRC 1694 | F. oxysporum | Oxysporum | Unknown | Human | MH582350.1 | [72] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Troni, E.; Beccari, G.; D’Amato, R.; Tini, F.; Baldo, D.; Senatore, M.T.; Beone, G.M.; Fontanella, M.C.; Prodi, A.; Businelli, D.; et al. In Vitro Evaluation of the Inhibitory Activity of Different Selenium Chemical Forms on the Growth of a Fusarium proliferatum Strain Isolated from Rice Seedlings. Plants 2021, 10, 1725. https://doi.org/10.3390/plants10081725
Troni E, Beccari G, D’Amato R, Tini F, Baldo D, Senatore MT, Beone GM, Fontanella MC, Prodi A, Businelli D, et al. In Vitro Evaluation of the Inhibitory Activity of Different Selenium Chemical Forms on the Growth of a Fusarium proliferatum Strain Isolated from Rice Seedlings. Plants. 2021; 10(8):1725. https://doi.org/10.3390/plants10081725
Chicago/Turabian StyleTroni, Elisabetta, Giovanni Beccari, Roberto D’Amato, Francesco Tini, David Baldo, Maria Teresa Senatore, Gian Maria Beone, Maria Chiara Fontanella, Antonio Prodi, Daniela Businelli, and et al. 2021. "In Vitro Evaluation of the Inhibitory Activity of Different Selenium Chemical Forms on the Growth of a Fusarium proliferatum Strain Isolated from Rice Seedlings" Plants 10, no. 8: 1725. https://doi.org/10.3390/plants10081725