Mice Mutated in the Third Fibronectin Domain of L1 Show Enhanced Hippocampal Neuronal Cell Death, Astrogliosis and Alterations in Behavior
Abstract
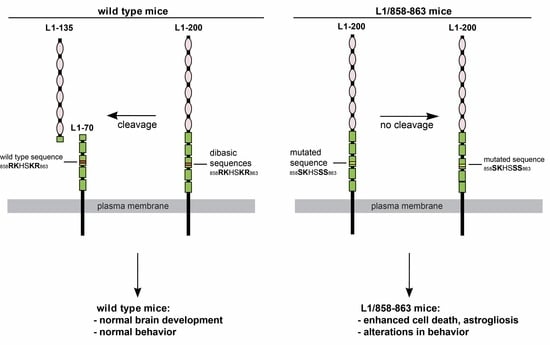
:1. Introduction
2. Materials and Methods
2.1. Mice
2.2. Reagents
2.3. Histology
2.4. Immunohistochemistry
2.5. Stereological Analyses
2.6. Behavior
2.6.1. Open Field and Elevated plus Maze
2.6.2. Social Interaction
2.6.3. Circadian Activity
2.6.4. Rotarod Test
2.6.5. Beam Walking
2.6.6. Grip Strength
2.6.7. Pole Test
2.6.8. Marble Burying
2.7. Statistics
3. Results
3.1. Behavioral Analysis
3.1.1. Body Weight and Grip Strength
3.1.2. Open Field
3.1.3. Elevated plus Maze
3.1.4. Social Interaction
3.1.5. Circadian Activity
3.1.6. Accelerating Rotarod and Pole Tests
3.1.7. Marble Burying
3.2. Histological Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Maness, P.F.; Schachner, M. Neural recognition molecules of the immunoglobulin superfamily: Signaling transducers of axon guidance and neuronal migration. Nat. Neurosci. 2007, 10, 19–26. [Google Scholar] [CrossRef]
- Sytnyk, V.; Leshchyns’ka, I.; Schachner, M. Neural Cell Adhesion Molecules of the Immunoglobulin Superfamily Regulate Synapse Formation, Maintenance, and Function. Trends Neurosci. 2017, 40, 295–308. [Google Scholar] [CrossRef]
- Kenwrick, S.; Watkins, A.; De Angelis, E. Neural cell recognition molecule L1: Relating biological complexity to human disease mutations. Hum. Mol. Genet. 2000, 9, 879–886. [Google Scholar] [CrossRef]
- Pollerberg, G.; Thelen, K.; Theiss, M.; Hochlehnert, B. The role of cell adhesion molecules for navigating axons: Density matters. Mech. Dev. 2013, 130, 359–372. [Google Scholar] [CrossRef]
- Stallcup, W.B.; Beasley, L. Involvement of the nerve growth factor-inducible large external glycoprotein (NILE) in neurite fasciculation in primary cultures of rat brain. Proc. Natl. Acad. Sci. USA 1985, 82, 1276–1280. [Google Scholar] [CrossRef]
- Lagenaur, C.; Lemmon, V. An L1-like molecule, the 8D9 antigen, is a potent substrate for neurite extension. Proc. Natl. Acad. Sci. USA 1987, 84, 7753–7757. [Google Scholar] [CrossRef]
- Grońska-Pęski, M.; Schachner, M.; Hébert, J.M. L1cam curbs the differentiation of adult-born hippocampal neurons. Stem Cell Res. 2020, 48, 101999. [Google Scholar] [CrossRef]
- Tonosaki, M.; Itoh, K.; Umekage, M.; Kishimoto, T.; Yaoi, T.; Lemmon, V.P.; Fushiki, S. L1cam Is Crucial for Cell Locomotion and Terminal Translocation of the Soma in Radial Migration during Murine Corticogenesis. PLoS ONE 2014, 9, e86186. [Google Scholar] [CrossRef]
- Weidle, U.H.; Eggle, D.; Klostermann, S. L1-CAM as a target for treatment of cancer with monoclonal antibodies. Anticancer Res. 2009, 29, 4919–4931. [Google Scholar]
- Godenschwege, T.A.; Kristiansen, L.V.; Uthaman, S.B.; Hortsch, M.; Murphey, R.K. A Conserved Role for Drosophila Neuroglian and Human L1-CAM in Central-Synapse Formation. Curr. Biol. 2006, 16, 12–23. [Google Scholar] [CrossRef]
- Wolfer, D.P.; Mohajeri, H.M.; Lipp, H.; Schachner, M. Increased flexibility and selectivity in spatial learning of transgenic mice ectopically expressing the neural cell adhesion molecule L1 in astrocytes. Eur. J. Neurosci. 1998, 10, 708–717. [Google Scholar] [CrossRef]
- Kraus, K.; Kleene, R.; Henis, M.; Braren, I.; Kataria, H.; Sharaf, A.; Loers, G.; Schachner, M.; Lutz, D. A Fragment of Adhesion Molecule L1 Binds to Nuclear Receptors to Regulate Synaptic Plasticity and Motor Coordination. Mol. Neurobiol. 2018, 55, 7164–7178. [Google Scholar] [CrossRef]
- Venero, C.; Tilling, T.; Hermans-Borgmeyer, I.; Herrero, A.I.; Schachner, M.; Sandi, C. Water maze learning and forebrain mRNA expression of the neural cell adhesion molecule L1. J. Neurosci. Res. 2004, 75, 172–181. [Google Scholar] [CrossRef]
- Silletti, S.; Mei, F.; Sheppard, D.; Montgomery, A.M. Plasmin-Sensitive Dibasic Sequences in the Third Fibronectin-like Domain of L1–Cell Adhesion Molecule (Cam) Facilitate Homomultimerization and Concomitant Integrin Recruitment. J. Cell Biol. 2000, 149, 1485–1502. [Google Scholar] [CrossRef]
- Kalus, I.; Schnegelsberg, B.; Seidah, N.G.; Kleene, R.; Schachner, M. The Proprotein Convertase PC5A and a Metalloprotease Are Involved in the Proteolytic Processing of the Neural Adhesion Molecule L1. J. Biol. Chem. 2003, 278, 10381–10388. [Google Scholar] [CrossRef]
- Lutz, D.; Wolters-Eisfeld, G.; Schachner, M.; Kleene, R. Cathepsin E generates a sumoylated intracellular fragment of the cell adhesion molecule L1 to promote neuronal and Schwann cell migration as well as myelination. J. Neurochem. 2014, 128, 713–724. [Google Scholar] [CrossRef]
- Lutz, D.; Loers, G.; Kleene, R.; Oezen, I.; Kataria, H.; Katagihallimath, N.; Braren, I.; Harauz, G.; Schachner, M. Myelin Basic Protein Cleaves Cell Adhesion Molecule L1 and Promotes Neuritogenesis and Cell Survival. J. Biol. Chem. 2014, 289, 13503–13518. [Google Scholar] [CrossRef]
- Linneberg, C.; Toft, C.L.F.; Kjaer-Sorensen, K.; Laursen, L.S. L1cam-mediated developmental processes of the nervous system are differentially regulated by proteolytic processing. Sci. Rep. 2019, 9, 3716. [Google Scholar] [CrossRef]
- Mechtersheimer, S.; Gutwein, P.; Agmon-Levin, N.; Stoeck, A.; Oleszewski, M.; Riedle, S.; Postina, R.; Fahrenholz, F.; Fogel, M.; Lemmon, V.; et al. Ectodomain shedding of L1 adhesion molecule promotes cell migration by autocrine binding to integrins. J. Cell Biol. 2001, 155, 661–674. [Google Scholar] [CrossRef]
- Lutz, D.; Kataria, H.; Kleene, R.; Loers, G.; Chaudhary, H.; Guseva, D.; Wu, B.; Jakovcevski, I.; Schachner, M. Myelin Basic Protein Cleaves Cell Adhesion Molecule L1 and Improves Regeneration After Injury. Mol. Neurobiol. 2016, 53, 3360–3376. [Google Scholar] [CrossRef]
- Kiefel, H.; Bondong, S.; Hazin, J.; Ridinger, J.; Schirmer, U.; Riedle, S.; Altevogt, P. L1CAM: A major driver for tumor cell invasion and motility. Cell Adhes. Migr. 2012, 6, 374–384. [Google Scholar] [CrossRef]
- Gavert, N.; Ben-Shmuel, A.; Lemmon, V.; Brabletz, T.; Ben-Ze’ev, A. Nuclear factor-kappaB signaling and ezrin are essential for L1-mediated metastasis of colon cancer cells. J. Cell Sci. 2010, 123, 2135–2143. [Google Scholar] [CrossRef]
- Schäfer, M.K.E.; Altevogt, P. L1CAM malfunction in the nervous system and human carcinomas. Cell. Mol. Life Sci. 2010, 67, 2425–2437. [Google Scholar] [CrossRef]
- Stoyanova, I.I.; Lutz, D. Functional diversity of neuronal cell adhesion and recognition molecule L1CAM through proteolytic cleavage. Cells 2022, 11, 3085. [Google Scholar] [CrossRef]
- Kraus, K.; Kleene, R.; Braren, I.; Loers, G.; Lutz, D.; Schachner, M. A fragment of adhesion molecule L1 is imported into mitochondria and regulates mitochondrial metabolism and trafficking. J. Cell Sci. 2018, 131, jcs210500. [Google Scholar] [CrossRef]
- Nagaraj, K.; Mualla, R.; Hortsch, M. The L1 Family of Cell Adhesion Molecules: A Sickening Number of Mutations and Protein Functions. Cell Adhes. Mol. Implic. Neurol. Dis. 2013, 8, 195–229. [Google Scholar] [CrossRef]
- Stumpel, C.; Vos, Y.J. L1 Syndrome. In GeneReviews((R)); Adam, M.P., Ardinger, H.H., Pagon, R.A., Wallace, S.E., Bean, L.J.H., Stephens, K., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 2004. [Google Scholar]
- Dahme, M.; Bartsch, U.; Martini, R.; Anliker, B.; Schachner, M.; Mantei, N. Disruption of the mouse L1 gene leads to malformations of the nervous system. Nat. Genet. 1997, 17, 346–349. [Google Scholar] [CrossRef]
- Loers, G.; Appel, D.; Lutz, D.; Congiu, L.; Kleene, R.; Hermans-Borgmeyer, I.; Schäfer, M.K.E.; Schachner, M. Amelioration of the abnormal phenotype of a new L1 syndrome mouse mutation with L1 mimetics. FASEB J. 2021, 35, e21329. [Google Scholar] [CrossRef]
- Rolf, B.; Kutsche, M.; Bartsch, U. Severe hydrocephalus in L1-deficient mice. Brain Res. 2001, 891, 247–252. [Google Scholar] [CrossRef]
- Rünker, A.E.; Bartsch, U.; Nave, K.A.; Schachner, M. The C264Y missense mutation in the extracellular domain of L1 impairs protein trafficking in vitro and in vivo. J. Neurosci. 2003, 23, 277–286. [Google Scholar] [CrossRef]
- Zhang, R.; Sun, X.-Z.; Cui, C.; Sakata-Haga, H.; Sawada, K.; Ye, C.; Fukui, Y. Spatial learning and expression of neural cell adhesion molecule L1 in rats X-irradiated prenatally. J. Med. Investig. 2007, 54, 322–330. [Google Scholar] [CrossRef] [PubMed]
- Sauce, B.; Wass, C.; Netrakanti, M.; Saylor, J.; Schachner, M.; Matzel, L.D. Heterozygous L1-deficient mice express an autism-like phenotype. Behav. Brain Res. 2015, 292, 432–442. [Google Scholar] [CrossRef] [PubMed]
- Kleene, R.; Lutz, D.; Loers, G.; Bork, U.; Borgmeyer, U.; Hermans-Borgmeyer, I.; Schachner, M. Revisiting the proteolytic processing of cell adhesion molecule L1. J. Neurochem. 2020, 157, 1102–1117. [Google Scholar] [CrossRef] [PubMed]
- Kilkenny, C.; Browne, W.J.; Cuthill, I.C.; Emerson, M.; Altman, D.G. Improving bioscience research reporting: The ARRIVE guidelines for reporting animal research. PLoS Biol. 2010, 8, e1000412. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Irintchev, A.; Rollenhagen, A.; Troncoso, E.; Kiss, J.Z.; Schachner, M. Structural and Functional Aberrations in the Cerebral Cortex of Tenascin-C Deficient Mice. Cereb. Cortex 2004, 15, 950–962. [Google Scholar] [CrossRef]
- Freitag, S.; Schachner, M.; Morellini, F. Behavioral alterations in mice deficient for the extracellular matrix glycoprotein tenascin-R. Behav. Brain Res. 2003, 145, 189–207. [Google Scholar] [CrossRef] [PubMed]
- Carola, V.; D’Olimpio, F.; Brunamonti, E.; Mangia, F.; Renzi, P. Evaluation of the elevated plus-maze and open-field tests for the assessment of anxiety-related behaviour in inbred mice. Behav. Brain Res. 2002, 134, 49–57. [Google Scholar] [CrossRef]
- Dunham, N.; Miya, T. A Note on a Simple Apparatus for Detecting Neurological Deficit in Rats and Mice**College of Pharmacy, University of Nebraska, Lincoln 8. J. Am. Pharm. Assoc. 1957, 46, 208–209. [Google Scholar] [CrossRef]
- Jones, B.J.; Roberts, D.J. The quantiative measurement of motor inco-ordination in naive mice using an acelerating rotarod. J. Pharm. Pharmacol. 1968, 20, 302–304. [Google Scholar] [CrossRef]
- Mehanna, A.; Mishra, B.; Kurschat, N.; Schulze, C.; Bian, S.; Loers, G.; Irintchev, A.; Schachner, M. Polysialic acid glycomimetics promote myelination and functional recovery after peripheral nerve injury in mice. Brain 2009, 132, 1449–1462. [Google Scholar] [CrossRef] [PubMed]
- Morellini, F.; Schachner, M. Enhanced novelty-induced activity, reduced anxiety, delayed resynchronization to daylight reversal and weaker muscle strength in tenascin-C-deficient mice. Eur. J. Neurosci. 2006, 23, 1255–1268. [Google Scholar] [CrossRef] [PubMed]
- Angoa-Perez, M.; Kane, M.J.; Briggs, D.I.; Francescutti, D.M.; Kuhn, D.M. Marble burying and nestlet shredding as tests of repetitive, compulsive-like behaviors in mice. J. Vis. Exp. 2013, 82, 50978. [Google Scholar] [CrossRef]
- De Boer, S.F.; Koolhaas, J.M. Defensive burying in rodents: Ethology, neurobiology and psychopharmacology. Eur. J. Pharmacol. 2003, 463, 145–161. [Google Scholar] [CrossRef] [PubMed]
- Schmid, J.S.; Bernreuther, C.; Nikonenko, A.G.; Ling, Z.; Mies, G.; Hossmann, K.-A.; Jakovcevski, I.; Schachner, M. Heterozygosity for the mutated X-chromosome-linked L1 cell adhesion molecule gene leads to increased numbers of neurons and enhanced metabolism in the forebrain of female carrier mice. Anat. Embryol. 2012, 218, 1375–1390. [Google Scholar] [CrossRef]
- Loers, G.; Kleene, R.; Minguez, M.G.; Schachner, M. The Cell Adhesion Molecule L1 Interacts with Methyl CpG Binding Protein 2 via Its Intracellular Domain. Int. J. Mol. Sci. 2022, 23, 3554. [Google Scholar] [CrossRef]
- Good, K.V.; Vincent, J.B.; Ausió, J. MeCP2: The Genetic Driver of Rett Syndrome Epigenetics. Front. Genet. 2021, 12, 620859. [Google Scholar] [CrossRef]
- Cohen, N.; Taylor, J.; Scott, L.; Guillery, R.; Soriano, P.; Furley, A. Errors in corticospinal axon guidance in mice lacking the neural cell adhesion molecule L1. Curr. Biol. 1998, 8, 26–33. [Google Scholar] [CrossRef]
- Fransen, E.; D’Hooge, R.; Van Camp, G.; Verhoye, M.; Sijbers, J.; Reyniers, E.; Soriano, P.; Kamiguchi, H.; Willemsen, R.; Koekkoek, S.K.; et al. L1 knockout mice show dilated ventricles, vermis hypoplasia and impaired exploration patterns. Hum. Mol. Genet. 1998, 7, 999–1009. [Google Scholar] [CrossRef]
- Schmalbach, B.; Lepsveridze, E.; Djogo, N.; Papashvili, G.; Kuang, F.; Leshchyns’Ka, I.; Sytnyk, V.; Nikonenko, A.G.; Dityatev, A.; Jakovcevski, I.; et al. Age-dependent loss of parvalbumin-expressing hippocampal interneurons in mice deficient in CHL1, a mental retardation and schizophrenia susceptibility gene. J. Neurochem. 2015, 135, 830–844. [Google Scholar] [CrossRef]
- Fransen, E.; Van Camp, G.; D’Hooge, R.; Vits, L.; Willems, P.J. Genotype-phenotype correlation in L1 associated diseases. J. Med. Genet. 1998, 35, 399–404. [Google Scholar] [CrossRef] [PubMed]
- Jakeman, L.B.; Chen, Y.; Lucin, K.M.; McTigue, D.M. Mice lacking L1 cell adhesion molecule have deficits in locomotion and exhibit enhanced corticospinal tract sprouting following mild contusion injury to the spinal cord. Eur. J. Neurosci. 2006, 23, 1997–2011. [Google Scholar] [CrossRef]
- Law, J.W.S.; Lee, A.Y.W.; Sun, M.; Nikonenko, A.G.; Chung, S.K.; Dityatev, A.; Schachner, M.; Morellini, F. Decreased Anxiety, Altered Place Learning, and Increased CA1 Basal Excitatory Synaptic Transmission in Mice with Conditional Ablation of the Neural Cell Adhesion Molecule L1. J. Neurosci. 2003, 23, 10419–10432. [Google Scholar] [CrossRef] [PubMed]
- Congiu, L.; Granato, V.; Loers, G.; Kleene, R.; Schachner, M. Mitochondrial and Neuronal Dysfunctions in L1 Mutant Mice. Int. J. Mol. Sci. 2022, 23, 4337. [Google Scholar] [CrossRef] [PubMed]
- Loers, G.; Kleene, R.; Bork, U.; Schachner, M. The interactions of the 70 kDa fragment of cell adhesion molecule L1 with topoisomerase 1, peroxisome proliferator-activated receptor gamma and NADH dehydrogenase (ubiquinone) flavoprotein 2 are involved in gene expression and neuronal L1-dependent functions. Int. J. Mol. Sci. 2023, 24, 2097. [Google Scholar] [PubMed]
- de Brouwer, G.; Fick, A.; Harvey, B.H.; Wolmarans, W. A critical inquiry into marble-burying as a preclinical screening paradigm of relevance for anxiety and obsessive-compulsive disorder: Mapping the way forward. Cogn. Affect. Behav. Neurosci. 2019, 19, 1–39. [Google Scholar] [CrossRef]
- Dixit, P.V.; Sahu, R.; Mishra, D.K. Marble-burying behavior test as a murine model of compulsive-like behavior. J. Pharmacol. Toxicol. Methods 2020, 102, 106676. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.X.; Tari, P.K.; She, K.; Haas, K. Neurexin-Neuroligin Cell Adhesion Complexes Contribute to Synaptotropic Dendritogenesis via Growth Stabilization Mechanisms In Vivo. Neuron 2010, 67, 967–983. [Google Scholar] [CrossRef] [PubMed]
- Budreck, E.C.; Kwon, O.-B.; Jung, J.H.; Baudouin, S.; Thommen, A.; Kim, H.-S.; Fukazawa, Y.; Harada, H.; Tabuchi, K.; Shigemoto, R.; et al. Neuroligin-1 controls synaptic abundance of NMDA-type glutamate receptors through extracellular coupling. Proc. Natl. Acad. Sci. USA 2013, 110, 725–730. [Google Scholar] [CrossRef]
- Barrow, S.L.; Constable, J.R.; Clark, E.; El-Sabeawy, F.; McAllister, A.K.; Washbourne, P. Neuroligin1: A cell adhesion molecule that recruits PSD-95 and NMDA receptors by distinct mechanisms during synaptogenesis. Neural Dev. 2009, 4, 17. [Google Scholar] [CrossRef] [PubMed]
- An, J.Y.; Cristino, A.S.; Zhao, Q.; Edson, J.; Williams, S.M.; Ravine, D.; Wray, J.; Marshall, V.M.; Hunt, A.; O Whitehouse, A.J.; et al. Towards a molecular characterization of autism spectrum disorders: An exome sequencing and systems approach. Transl. Psychiatry 2014, 4, e394. [Google Scholar] [CrossRef] [PubMed]
- Espinosa, F.; Xuan, Z.; Liu, S.; Powell, C.M. Neuroligin 1 modulates striatal glutamatergic neurotransmission in a pathway and NMDAR subunit-specific manner. Front. Synaptic Neurosci. 2015, 7, 11. [Google Scholar] [CrossRef] [PubMed]
- Trobiani, L.; Meringolo, M.; Diamanti, T.; Bourne, Y.; Marchot, P.; Martella, G.; Dini, L.; Pisani, A.; De Jaco, A.; Bonsi, P. The neuroligins and the synaptic pathway in Autism Spectrum Disorder. Neurosci. Biobehav. Rev. 2020, 119, 37–51. [Google Scholar] [CrossRef] [PubMed]
- Appel, F.; Holm, J.; Conscience, J.; Schachner, M. Several extracellular domains of the neural cell adhesion molecule L1 are involved in neurite outgrowth and cell body adhesion. J. Neurosci. 1993, 13, 4764–4775. [Google Scholar] [CrossRef] [PubMed]
- Holm, J.; Appel, F.; Schachner, M. Several extracellular domains of the neural cell adhesion molecule L1 are involved in homophilic interactions. J. Neurosci. Res. 1995, 42, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Stallcup, W.B. The third fibronectin type III repeat is required for L1 to serve as an optimal substratum for neurite extension. J. Neurosci. Res. 2000, 61, 33–43. [Google Scholar] [CrossRef]
- Nayeem, N.; Silletti, S.; Yang, X.; Lemmon, V.P.; A Reisfeld, R.; Stallcup, W.B.; Montgomery, A.M. A potential role for the plasmin(ogen) system in the posttranslational cleavage of the neural cell adhesion molecule L1. J. Cell Sci. 1999, 112, 4739–4749. [Google Scholar] [CrossRef]
- Guo, D.; Shi, Y.; Jian, W.; Fu, Y.; Yang, H.; Guo, M.; Yong, W.; Chen, G.; Deng, H.; Qin, Y.; et al. A novel nonsense mutation in the L1CAM gene responsible for X-linked congenital hydrocephalus. J. Gene Med. 2020, 22, e3180. [Google Scholar] [CrossRef]
- Shaw, M.; Yap, T.Y.; Henden, L.; Bahlo, M.; Gardner, A.; Kalscheuer, V.M.; Haan, E.; Christie, L.; Hackett, A.; Gecz, J. Identical by descent L1CAM mutation in two apparently unrelated families with intellectual disability without L1 syndrome. Eur. J. Med. Genet. 2015, 58, 364–368. [Google Scholar] [CrossRef]
- Vos, Y.J.; Hofstra, R.M.W. An updated and upgraded L1CAM mutation database. Hum. Mutat. 2010, 31, E1102–E1109. [Google Scholar] [CrossRef]
- Finckh, U.; Ressler, B.; Veske, A.; Gal, A. Spectrum and detection rate ofL1CAM mutations in isolated and familial cases with clinically suspected L1-disease. Am. J. Med. Genet. 2000, 92, 40–46. [Google Scholar] [CrossRef]
- Kanemura, Y.; Okamoto, N.; Sakamoto, H.; Shofuda, T.; Kamiguchi, H.; Yamasaki, M. Molecular mechanisms and neuroimaging criteria for severe L1 syndrome with X-linked hydrocephalus. J. Neurosurg. Pediatr. 2006, 105, 403–412. [Google Scholar] [CrossRef] [PubMed]
- Sato, M.S.; Kyriakopoulos, M.; James, A.; Marwedel, S.; Borsay, C.; Gutierrez, A.A.; Blakemore, A.I.; Need, A.C. Hemizygous mutations in L1CAM in two unrelated male probands with childhood onset psychosis. Psychiatr. Genet. 2020, 30, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Saugier-Veber, P.; Martin, C.; Le Meur, N.; Lyonnet, S.; Munnich, A.; David, A.; Hénocq, A.; Héron, D.; Jonveaux, P.; Odent, S.; et al. Identification of novelL1CAM mutations using fluorescence-assisted mismatch analysis. Hum. Mutat. 1998, 12, 259–266. [Google Scholar] [CrossRef]
- Ardalan, M.; Chumak, T.; Quist, A.; Hermans, E.; Rafati, A.H.; Gravina, G.; Shiadeh, S.M.J.; Svedin, P.; Alabaf, S.; Hansen, B.; et al. Reelin cells and sex-dependent synaptopathology in autism following postnatal immune activation. Br. J. Pharmacol. 2022, 179, 4400–4422. [Google Scholar] [CrossRef]
- Zhang, R.; Wu, L.; Eckert, T.; Burg-Roderfeld, M.; Rojas-Macias, M.A.; Lutteke, T.; Krylov, V.B.; Argunov, D.A.; Datta, A.; Markart, P.; et al. Lysozyme’s lectin-like characteristics facilitates its immune defense function. Q Rev. Biophys. 2017, 50, e9. [Google Scholar] [CrossRef]
- Luders, E.; Kurth, F.; Das, D.; Oyarce, D.E.; Shaw, M.E.; Sachdev, P.; Easteal, S.; Anstey, K.J.; Cherbuin, N. Associations between corpus callosum size and ADHD symptoms in older adults: The PATH through life study. Psychiatry Res. Neuroimaging 2016, 256, 8–14. [Google Scholar] [CrossRef]
- Itoh, K.; Cheng, L.; Kamei, Y.; Fushiki, S.; Kamiguchi, H.; Gutwein, P.; Stoeck, A.; Arnold, B.; Altevogt, P.; Lemmon, V. Brain development in mice lacking L1–L1 homophilic adhesion. J. Cell Biol. 2004, 165, 145–154. [Google Scholar] [CrossRef]
- Arami, S.; Jucker, M.; Schachner, M.; Welzl, H. The effect of continuous intraventricular infusion of L1 and NCAM antibodies on spatial learning in rats. Behav. Brain Res. 1996, 81, 81–87. [Google Scholar] [CrossRef]
- Tsoory, M.M.; Guterman, A.; Richter-Levin, G. “Juvenile stress” alters maturation-related changes in expression of the neural cell adhesion molecule L1 in the limbic system: Relevance for stress-related psychopathologies. J. Neurosci. Res. 2010, 88, 369–380. [Google Scholar] [CrossRef]
- Demyanenko, G.P.; Tsai, A.Y.; Maness, P.F. Abnormalities in Neuronal Process Extension, Hippocampal Development, and the Ventricular System of L1 Knockout Mice. J. Neurosci. 1999, 19, 4907–4920. [Google Scholar] [CrossRef] [PubMed]
- Johri, A.; Beal, M.F. Mitochondrial Dysfunction in Neurodegenerative Diseases. Experiment 2012, 342, 619–630. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, E.; Musich, P.R.; Lin, F. Mitochondrial dysfunction in neurodegenerative diseases and the potential countermeasure. CNS Neurosci. Ther. 2019, 25, 816–824. [Google Scholar] [CrossRef] [PubMed]
- Picard, M.; McEwen, B.S. Psychological Stress and Mitochondria: A Conceptual Framework. Psychosom. Med. 2018, 80, 126–140. [Google Scholar] [CrossRef] [PubMed]









Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Congiu, L.; Granato, V.; Jakovcevski, I.; Kleene, R.; Fernandes, L.; Freitag, S.; Kneussel, M.; Schachner, M.; Loers, G. Mice Mutated in the Third Fibronectin Domain of L1 Show Enhanced Hippocampal Neuronal Cell Death, Astrogliosis and Alterations in Behavior. Biomolecules 2023, 13, 776. https://doi.org/10.3390/biom13050776
Congiu L, Granato V, Jakovcevski I, Kleene R, Fernandes L, Freitag S, Kneussel M, Schachner M, Loers G. Mice Mutated in the Third Fibronectin Domain of L1 Show Enhanced Hippocampal Neuronal Cell Death, Astrogliosis and Alterations in Behavior. Biomolecules. 2023; 13(5):776. https://doi.org/10.3390/biom13050776
Chicago/Turabian StyleCongiu, Ludovica, Viviana Granato, Igor Jakovcevski, Ralf Kleene, Luciana Fernandes, Sandra Freitag, Matthias Kneussel, Melitta Schachner, and Gabriele Loers. 2023. "Mice Mutated in the Third Fibronectin Domain of L1 Show Enhanced Hippocampal Neuronal Cell Death, Astrogliosis and Alterations in Behavior" Biomolecules 13, no. 5: 776. https://doi.org/10.3390/biom13050776