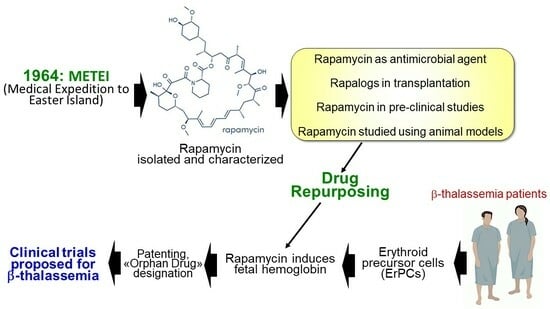
The Long Scientific Journey of Sirolimus (Rapamycin): From the Soil of Easter Island (Rapa Nui) to Applied Research and Clinical Trials on β-Thalassemia and Other Hemoglobinopathies
Abstract
:Simple Summary
Abstract
1. Introduction
2. From the METEI Expedition to the Discovery of Rapamycin (Sirolimus)
3. Rapamycin: Biomedical Applications
4. Rapamycin in Organ and Tissue Transplantation
5. Rapamycin for Longevity?
6. Rapamycin: Effects on the Immune System
7. Testing Rapamycin (Sirolimus) on Erythroid Cells: Induction of Fetal Hemoglobin
8. Testing Rapamycin on Animal Models: Supporting Evidences for a Possible Role in the Therapy of β-Thalassemia
9. IP Protection and Orphan Drug Designation
10. mTOR Inhibitors and Clinical Case Reports: First Evidences on In Vivo Effects on HbF
11. The Sirthalaclin and Thala-Rap Clinical Trials on β-Thalassemia
12. Rapamycin and Biomedical Applications in Hematology: The Journey Is Not over Yet
13. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Weatherall, D.J. Phenotype-genotype relationships in monogenic disease: Lessons from the thalassaemias. Nat. Rev. Genet. 2001, 2, 245–255. [Google Scholar] [CrossRef]
- Origa, R. β-Thalassemia. Genet. Med. 2017, 19, 609–619. [Google Scholar] [CrossRef] [PubMed]
- Fucharoen, S.; Weatherall, D.J. Progress toward the Control and Management of the Thalassemias. Hematol. Oncol. Clin. N. Am. 2016, 30, 359–371. [Google Scholar] [CrossRef]
- Sripichai, O.; Fucharoen, S. Fetal hemoglobin regulation in β-thalassemia: Heterogeneity, modifiers and therapeutic approaches. Expert Rev. Hematol. 2016, 9, 1129–1137. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Zhang, X.; Yu, L.; Cai, R.; Ma, X.; Zheng, C.; Zhou, Y.; Liu, Q.; Wei, X.; Lin, L.; et al. KLF1 mutations are relatively more common in a thalassemia endemic region and ameliorate the severity of β-thalassemia. Blood 2014, 124, 803–811. [Google Scholar] [CrossRef] [PubMed]
- Musallam, K.M.; Sankaran, V.G.; Cappellini, M.D.; Duca, L.; Nathan, D.G.; Taher, A.T. Fetal hemoglobin levels and morbidity in untransfused patients with β-thalassemia intermedia. Blood 2012, 119, 364–367. [Google Scholar] [CrossRef] [PubMed]
- Forget, B.G. Molecular basis of hereditary persistence of fetal hemoglobin. Ann. N. Y. Acad. Sci. 1998, 850, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Nuinoon, M.; Makarasara, W.; Mushiroda, T.; Setianingsih, I.; Wahidiyat, P.A.; Sripichai, O.; Kumasaka, N.; Takahashi, A.; Svasti, S.; Munkongdee, T.; et al. A genome-wide association identified the common genetic variants influence disease severity in β0-thalassemia/hemoglobin E. Hum. Genet. 2010, 127, 303–314. [Google Scholar] [CrossRef]
- Galanello, R.; Sanna, S.; Perseu, L.; Sollaino, M.C.; Satta, S.; Lai, M.E.; Barella, S.; Uda, M.; Usala, G.; Abecasis, G.R.; et al. Amelioration of Sardinian β0 thalassemia by genetic modifiers. Blood 2009, 114, 3935–3937. [Google Scholar] [CrossRef]
- Danjou, F.; Anni, F.; Perseu, L.; Satta, S.; Dessì, C.; Lai, M.E.; Fortina, P.; Devoto, M.; Galanello, R. Genetic modifiers of b-thalassemia and clinical severity as assessed by age at first transfusion. Haematologica 2012, 97, 989–993. [Google Scholar] [CrossRef]
- Badens, C.; Joly, P.; Agouti, I.; Thuret, I.; Gonnet, K.; Fattoum, S.; Francina, A.; Simeoni, M.C.; Loundou, A.; Pissard, S. Variants in genetic modifiers of β-thalassemia can help to predict the major or intermedia type of the disease. Haematologica 2011, 96, 1712–1714. [Google Scholar] [CrossRef] [PubMed]
- Uda, M.; Galanello, R.; Sanna, S.; Lettre, G.; Sankaran, V.G.; Chen, W.; Usala, G.; Busonero, F.; Maschio, A.; Albai, G.; et al. Genome wide association study shows BCL11A associated with persistent fetal hemoglobin and amelioration of the phenotype of β-thalassemia. Proc. Natl. Acad. Sci. USA 2008, 105, 1620–1625. [Google Scholar] [CrossRef] [PubMed]
- Halford, B. Rapamycin’s secrets unearthed. Chem. Eng. News 2016, 94, 26–30. [Google Scholar]
- Boutilier, J.A. METEI: A Canadian medical expedition to Easter Island, 1964–1965. Rapa Nui J. 1992, 6, 21–34. [Google Scholar]
- Hobby, G.; Clark, R.; Woywodt, A. A treasure from a barren island: The discovery of rapamycin. Clin. Kidney J. 2022, 15, 1971–1972. [Google Scholar] [CrossRef] [PubMed]
- Sehgal, S.N.; Baker, H.; Vézina, C. Rapamycin (AY-22,989), a new antifungal antibiotic. II. Fermentation, isolation and characterization. J. Antibiot. 1975, 28, 727–732. [Google Scholar] [CrossRef] [PubMed]
- Vézina, C.; Kudelski, A.; Sehgal, S.N. Rapamycin (AY-22,989), a new antifungal antibiotic. I. Taxonomy of the producing streptomycete and isolation of the active principle. J. Antibiot. 1975, 28, 721–726. [Google Scholar] [CrossRef]
- Douros, J.; Suffness, M. New antitumor substances of natural origin. Cancer Treat. Rev. 1981, 8, 63–87. [Google Scholar] [CrossRef]
- Seto, B. Rapamycin and mTOR: A serendipitous discovery and implications for breast cancer. Clin. Transl. Med. 2012, 1, 29. [Google Scholar] [CrossRef]
- Powers, T. The origin story of rapamycin: Systemic bias in biomedical research and cold war politics. Mol. Biol. Cell 2022, 33, pe7. [Google Scholar] [CrossRef]
- Kwon, M.; Han, J.; Kim, U.J.; Cha, M.; Um, S.W.; Bai, S.J.; Hong, S.K.; Lee, B.H. Inhibition of Mammalian Target of Rapamycin (mTOR) Signaling in the Insular Cortex Alleviates Neuropathic Pain after Peripheral Nerve Injury. Front. Mol. Neurosci. 2017, 10, 79. [Google Scholar] [CrossRef]
- Sun, S.Y.; Rosenberg, L.M.; Wang, X.; Zhou, Z.; Yue, P.; Fu, H. Khuri FR: Activation of Akt and eIF4E survival pathways by rapamycin-mediated mammalian target of rapamycin inhibition. Cancer Res. 2005, 65, 7052–7058. [Google Scholar] [CrossRef]
- Chan, S. Targeting the mammalian target of rapamycin (mTOR): A new approach to treating cancer. Br. J. Cancer 2004, 91, 1420–1424. [Google Scholar] [CrossRef] [PubMed]
- Sirolimus. Available online: https://go.drugbank.com/drugs/DB00877 (accessed on 3 July 2023).
- Saunders, R.N.; Metcalfe, M.S.; Nicholson, M.L. Rapamycin in transplantation: A review of the evidence. Kidney Int. 2001, 59, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Saxton, R.A.; Sabatini, D.M. mTOR Signaling in Growth, Metabolism, and Disease. Cell 2017, 169, 361–371. [Google Scholar] [CrossRef]
- Kahan, B.D. Sirolimus: A new agent for clinical renal transplantation. Transplant. Proc. 1997, 29, 48–50. [Google Scholar] [CrossRef] [PubMed]
- Vasquez, E.M. Sirolimus: A new agent for prevention of renal allograft rejection. Am. J. Health Syst. Pharm. 2000, 57, 437–448. [Google Scholar] [CrossRef] [PubMed]
- Hernández, D.; Martínez, D.; Gutiérrez, E.; López, V.; Gutiérrez, C.; García, P.; Cobelo, C.; Cabello, M.; Burgos, D.; Sola, E.; et al. Clinical evidence on the use of anti-mTOR drugs in renal transplantation. Nefrologia 2011, 31, 27–34. [Google Scholar]
- Schaffer, S.A.; Ross, H.J. Everolimus: Efficacy and safety in cardiac transplantation. Expert Opin. Drug Saf. 2010, 9, 843–854. [Google Scholar] [CrossRef]
- Tang, C.Y.; Shen, A.; Wei, X.F.; Li, Q.D.; Liu, R.; Deng, H.J.; Wu, Y.Z.; Wu, Z.J. Everolimus in de novo liver transplant recipients: A systematic review. Hepatobiliary Pancreat. Dis. Int. 2015, 14, 461–469. [Google Scholar] [CrossRef]
- Ji, L.; Xie, W.; Zhang, Z. Efficacy and safety of sirolimus in patients with systemic lupus erythematosus: A systematic review and meta-analysis. Semin. Arthritis Rheum. 2020, 50, 1073–1080. [Google Scholar] [CrossRef]
- Wang, Q.; Luo, M.; Xiang, B.; Chen, S.; Ji, Y. The efficacy and safety of pharmacological treatments for lymphangioleiomyomatosis. Respir. Res. 2020, 21, 55. [Google Scholar] [CrossRef]
- Sasongko, T.H.; Ismail, N.F.; Zabidi-Hussin, Z. Rapamycin and rapalogs for tuberous sclerosis complex. Cochrane Database Syst. Rev. 2016, 7, CD011272. [Google Scholar] [CrossRef]
- Graillon, T.; Sanson, M.; Campello, C.; Idbaih, A.; Peyre, M.; Peyrière, H.; Basset, N.; Autran, D.; Roche, C.; Kalamarides, M.; et al. Everolimus and Octreotide for Patients with Recurrent Meningioma: Results from the Phase II CEVOREM Trial. Clin. Cancer Res. 2020, 26, 552–557. [Google Scholar] [CrossRef]
- Gallo, M.; Malandrino, P.; Fanciulli, G.; Rota, F.; Faggiano, A.; Colao, A.; NIKE Group. Everolimus as first line therapy for pancreatic neuroendocrine tumours: Current knowledge and future perspectives. J. Cancer Res. Clin. Oncol. 2017, 143, 1209–1224. [Google Scholar] [CrossRef]
- Manohar, P.M.; Beesley, L.J.; Taylor, J.M.; Hesseltine, E.; Haymart, M.R.; Esfandiari, N.H.; Hanauer, D.A.; Worden, F.P. Retrospective Study of Sirolimus and Cyclophosphamide in Patients with Advanced Differentiated Thyroid Cancers. J. Thyroid. Disord. Ther. 2015, 4, 188. [Google Scholar] [CrossRef] [PubMed]
- Hortobagyi, G.N. Everolimus plus exemestane for the treatment of advanced breast cancer: A review of subanalyses from BOLERO-2. Neoplasia 2015, 17, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Merli, M.; Ferrario, A.; Maffioli, M.; Arcaini, L.; Passamonti, F. Everolimus in diffuse large B-cell lymphomas. Future Oncol. 2015, 11, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Motzer, R.J.; Escudier, B.; Oudard, S.; Hutson, T.E.; Porta, C.; Bracarda, S.; Grünwald, V.; Thompson, J.A.; Figlin, R.A.; Hollaender, N.; et al. Phase 3 trial of everolimus for metastatic renal cell carcinoma: Final results and analysis of prognostic factors. Cancer 2010, 116, 4256–4265. [Google Scholar] [CrossRef]
- Augustine, J.J.; Bodziak, K.A.; Hricik, D.E. Use of sirolimus in solid organ transplantation. Drugs 2007, 67, 369–391. [Google Scholar] [CrossRef]
- Hartinger, J.M.; Ryšánek, P.; Slanař, O.; Šíma, M. Pharmacokinetic principles of dose adjustment of mTOR inhibitors in solid organ transplanted patients. J. Clin. Pharm. Ther. 2022, 47, 1362–1367. [Google Scholar] [CrossRef]
- Ventura-Aguiar, P.; Campistol, J.M.; Diekmann, F. Safety of mTOR inhibitors in adult solid organ transplantation. Expert Opin. Drug Saf. 2016, 15, 303–319. [Google Scholar] [CrossRef]
- Toniato de Rezende Freschi, J.; Cristelli, M.P.; Viana, L.A.; Ficher, K.N.; Nakamura, M.R.; Proença, H.; Dreige, Y.C.; de Marco, R.; de Lima, M.G.; Foresto, R.D.; et al. A Head-to-head Comparison of De Novo Sirolimus or Everolimus Plus Reduced-dose Tacrolimus in Kidney Transplant Recipients: A Prospective and Randomized Trial. Transplantation 2023. [Google Scholar] [CrossRef]
- Tomita, Y.; Uehara, S.; Takiguchi, S.; Nakamura, M. Effect of Mammalian Target of Rapamycin Inhibition on Activated Regulatory T-Cell Expansion in Kidney Transplantation. Transplant. Proc. 2023, 55, 792–796. [Google Scholar] [CrossRef]
- Nunes Ficher, K.; Dreige, Y.; Gessolo Lins, P.R.; Nicolau Ferreira, A.; Toniato de Rezende Freschi, J.; Linhares, K.; Stopa Martins, S.; Custodio, L.; Cristelli, M.; Viana, L.; et al. Long-term Efficacy and Safety of Everolimus versus Mycophenolate in Kidney Transplant Recipients Receiving Tacrolimus. Transplantation 2022, 106, 381–390. [Google Scholar] [CrossRef]
- de Souza, A.R.; Dos Santos, T.A.G.M.; Von Jakitsch, C.B.; de Sant’Anna, A.L.G.G.; de Claudio, J.C.M.; Branco, J.N.R.; Giovanazzi, R.S.D.; Junior, N.A.H.; Pimentel, W.S.; da Costa, S.A.C.M.; et al. Mammalian Target of Rapamycin Inhibitors vs. Calcineurin Inhibitors in Chronic Graft Rejection after Lung Transplantation: A Systematic Review and Meta-Analysis. Transplant. Proc. 2021, 53, 3056–3064. [Google Scholar] [CrossRef]
- Jang, S.C.; Oh, B.C.; Nam, J.H.; Lee, E.K.; Kim, H.L.; Kwon, S.H. Clinical impact and economic burden of post-transplant infections following heart transplantation: A retrospective nationwide cohort study. J. Heart Lung Transplant. 2022, 41, 1601–1610. [Google Scholar] [CrossRef] [PubMed]
- Vidigal, A.C.; de Lucena, D.D.; Beyerstedt, S.; Rangel, É.B. A comprehensive update of the metabolic and toxicological considerations for immunosuppressive drugs used during pancreas transplantation. Expert Opin. Drug Metab. Toxicol. 2023, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Panackel, C.; Mathew, J.F.; Fawas, N.M.; Jacob, M. Immunosuppressive Drugs in Liver Transplant: An Insight. J. Clin. Exp. Hepatol. 2022, 12, 1557–1571. [Google Scholar] [CrossRef] [PubMed]
- Merola, J.; Shamim, A.; Weiner, J. Update on immunosuppressive strategies in intestinal transplantation. Curr. Opin. Organ. Transplant. 2022, 27, 119–125. [Google Scholar] [CrossRef]
- Li, X.; Chen, K.; Wang, Z.; Li, J.; Wang, X.; Xie, C.; Tong, J.; Shen, Y. The mTOR signalling in corneal diseases: A recent update. Biochem. Pharmacol. 2023, 213, 115620. [Google Scholar] [CrossRef] [PubMed]
- Giglio, F.; Xue, E.; Barone, A.; Lorentino, F.; Greco, R.; Ruggeri, A.; Zambelli, M.; Parisi, C.; Milani, R.; Clerici, D.; et al. Intrabone Transplantation of a Single Unwashed Umbilical Cord Blood Unit with Antithymocyte Globulin-Free and Sirolimus-Based Graft-versus-Host Disease Prophylaxis: Fast Immune Reconstitution and Long-Term Disease Control in Patients with High-Risk Diseases. Transplant. Cell. Ther. 2023, 29, 519.e1–519.e9. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, Y.; Zhou, L.; Du, G.S.; He, Q. Trends of rapamycin in survival benefits of liver transplantation for hepatocellular carcinoma. World J. Gastrointest. Surg. 2021, 13, 953–966. [Google Scholar] [CrossRef] [PubMed]
- Dahle, D.O.; Skauby, M.; Langberg, C.W.; Brabrand, K.; Wessel, N.; Midtvedt, K. Renal Cell Carcinoma and Kidney Transplantation: A Narrative Review. Transplantation 2022, 106, e52–e63. [Google Scholar] [CrossRef]
- Lazzari, L.; Balaguer-Roselló, A.; Montoro, J.; Greco, R.; Hernani, R.; Lupo-Stanghellini, M.T.; Villalba, M.; Giglio, F.; Facal, A.; Lorentino, F.; et al. Post-transplant cyclophosphamide and sirolimus based graft-versus-host disease prophylaxis after allogeneic stem cell transplantation for acute myeloid leukemia. Bone Marrow Transplant. 2022, 57, 1389–1398. [Google Scholar] [CrossRef] [PubMed]
- Coloni, G.F.; Venuta, F.; Ciccone, A.M.; Rendina, E.A.; De Giacomo, T.; Filice, M.J.; Diso, D.; Anile, M.; Andreetti, C.; Aratari, M.T.; et al. Lung transplantation for cystic fibrosis. Transplant. Proc. 2004, 36, 648–650. [Google Scholar] [CrossRef]
- Uygun, V.; Keleş, S.; Daloğlu, H.; Öztürkmen, S.; Yalçın, K.; Karasu, G.; Yeşilipek, A. Hematopoietic stem cell transplantation in serine/threonine kinase 4 (STK4) deficiency: Report of two cases and literature review. Pediatr. Transplant. 2023, 27, e14439. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, M.M.; Fitzhugh, C.D.; Weitzel, R.P.; Link, M.E.; Coles, W.A.; Zhao, X.; Rodgers, G.P.; Powell, J.D.; Tisdale, J.F. Nonmyeloablative HLA-matched sibling allogeneic hematopoietic stem cell transplantation for severe sickle cell phenotype. JAMA 2014, 312, 48–56. [Google Scholar] [CrossRef]
- Tedesco-Silva, H.; Saliba, F.; Barten, M.J.; De Simone, P.; Potena, L.; Gottlieb, J.; Gawai, A.; Bernhardt, P.; Pascual, J. An overview of the efficacy and safety of everolimus in adult solid organ transplant recipients. Transplant. Rev. 2022, 36, 100655. [Google Scholar] [CrossRef]
- Urzì Brancati, V.; Scarpignato, C.; Minutoli, L.; Pallio, G. Use of Pharmacogenetics to Optimize Immunosuppressant Therapy in Kidney-Transplanted Patients. Biomedicines 2022, 10, 1798. [Google Scholar] [CrossRef]
- Jou, S.; Mendez, S.R.; Feinman, J.; Mitrani, L.R.; Fuster, V.; Mangiola, M.; Moazami, N.; Gidea, C. Heart transplantation: Advances in expanding the donor pool and xenotransplantation. Nat. Rev. Cardiol. 2023. [Google Scholar] [CrossRef]
- Maenaka, A.; Kinoshita, K.; Hara, H.; Cooper, D.K.C. The case for the therapeutic use of mechanistic/mammalian target of rapamycin (mTOR) inhibitors in xenotransplantation. Xenotransplantation 2023, 30, e12802. [Google Scholar] [CrossRef]
- Vellai, T.; Takacs-Vellai, K.; Zhang, Y.; Kovacs, A.L.; Orosz, L.; Müller, F. Influence of TOR kinase on lifespan in C. elegans. Nature 2003, 426, 620. [Google Scholar] [CrossRef] [PubMed]
- Powers, R.W., 3rd; Kaeberlein, M.; Caldwell, S.D.; Kennedy, B.K.; Fields, S. Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev. 2006, 20, 174–184. [Google Scholar] [CrossRef]
- Kapahi, P.; Zid, B.M.; Harper, T.; Koslover, D.; Sapin, V.; Benzer, S. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr. Biol. 2004, 14, 885–890. [Google Scholar] [CrossRef] [PubMed]
- Harrison, D.E.; Strong, R.; Sharp, Z.D.; Nelson, J.F.; Astle, C.M.; Flurkey, K.; Nadon, N.L.; Wilkinson, J.E.; Frenkel, K.; Carter, C.S.; et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature 2009, 460, 392–395. [Google Scholar] [CrossRef]
- Johnson, S.C.; Rabinovitch, P.S.; Kaeberlein, M. mTOR is a key modulator of ageing and age-related disease. Nature 2013, 493, 338–345. [Google Scholar] [CrossRef]
- Kaeberlein, T.L.; Green, A.S.; Haddad, G.; Hudson, J.; Isman, A.; Nyquist, A.; Rosen, B.S.; Suh, Y.; Zalzala, S.; Zhang, X.; et al. Evaluation of off-label rapamycin use to promote healthspan in 333 adults. Geroscience 2023, 1–12. [Google Scholar] [CrossRef]
- Keating, R.; Hertz, T.; Wehenkel, M.; Harris, T.L.; Edwards, B.A.; McClaren, J.L.; Brown, S.A.; Surman, S.; Wilson, Z.S.; Bradley, P.; et al. The kinase mTOR modulates the antibody response to provide cross-protective immunity to lethal infection with influenza virus. Nat. Immunol. 2013, 14, 1266–1276. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J. Infectious disease. Immune suppressant unexpectedly boosts flu vaccine. Science 2013, 342, 413. [Google Scholar] [CrossRef] [PubMed]
- Frasca, D.; Blomberg, B.B. Inflammaging decreases adaptive and innate immune responses in mice and humans. Biogerontology 2016, 17, 7–19. [Google Scholar] [CrossRef]
- Bak, S.; Tischer, S.; Dragon, A.; Ravens, S.; Pape, L.; Koenecke, C.; Oelke, M.; Blasczyk, R.; Maecker-Kolhoff, B.; Eiz-Vesper, B. Selective Effects of mTOR Inhibitor Sirolimus on Naïve and CMV-Specific T Cells Extending Its Applicable Range Beyond Immunosuppression. Front. Immunol. 2018, 9, 2953. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.W.; Veitch, M.; Bridge, J.A.; Overgaard, N.H.; Cruz, J.L.; Linedale, R.; Franklin, M.E.; Saunders, N.A.; Simpson, F.; Frazer, I.H.; et al. Clinically-Relevant Rapamycin Treatment Regimens Enhance CD8+ Effector Memory T Cell Function in the Skin and Allow their Infiltration into Cutaneous Squamous Cell Carcinoma. Oncoimmunology 2018, 7, e1479627. [Google Scholar] [CrossRef]
- Amiel, E.; Everts, B.; Freitas, T.C.; King, I.L.; Curtis, J.D.; Pearce, E.L.; Pearce, E.J. Inhibition of mechanistic target of rapamycin promotes dendritic cell activation and enhances therapeutic autologous vaccination in mice. J. Immunol. 2012, 189, 2151–2158. [Google Scholar] [CrossRef]
- Araki, K.; Youngblood, B.; Ahmed, R. The role of mTOR in memory CD8 T-cell differentiation. Immunol. Rev. 2010, 235, 234–243. [Google Scholar] [CrossRef] [PubMed]
- Zurlo, M.; Nicoli, F.; Proietto, D.; Dallan, B.; Zuccato, C.; Cosenza, L.C.; Gasparello, J.; Papi, C.; d’Aversa, E.; Borgatti, M.; et al. Effects of Sirolimus treatment on patients with β-Thalassemia: Lymphocyte immunophenotype and biological activity of memory CD4+ and CD8+ T cells. J. Cell. Mol. Med. 2023, 27, 353–364. [Google Scholar] [CrossRef] [PubMed]
- Lozzio, B.B.; Lozzio, C.B. Properties of the K562 cell line derived from a patient with chronic myeloid leukemia. Int. J. Cancer 1977, 19, 136. [Google Scholar] [CrossRef]
- Andersson, L.C.; Jokinen, M.; Gahmberg, C.G. Induction of erythroid differentiation in the human leukaemia cell line K562. Nature 1979, 278, 364–365. [Google Scholar] [CrossRef]
- Gahmberg, C.G.; Jokinen, M.; Andersson, L.C. Expression of the major red cell sialoglycoprotein, glycophorin A, in the human leukemic cell line K562. J. Biol. Chem. 1979, 254, 7442–7448. [Google Scholar] [CrossRef]
- Villeval, J.L.; Pelicci, P.G.; Tabilio, A.; Titeux, M.; Henri, A.; Houesche, F.; Thomopoulos, P.; Vainchenker, W.; Garbaz, M.; Rochant, H.; et al. Erythroid properties of K562 cells. Effect of hemin, butyrate and TPA induction. Exp. Cell Res. 1983, 146, 428–435. [Google Scholar] [CrossRef]
- Rutherford, T.R.; Clegg, J.B.; Weatherall, D.J. K562 human leukaemic cells synthesise embryonic haemoglobin in response to haemin. Nature 1979, 280, 164–165. [Google Scholar] [CrossRef]
- Theodorou, A.; Phylactides, M.; Forti, L.; Cramarossa, M.R.; Spyrou, P.; Gambari, R.; Thein, S.L.; Kleanthous, M. The investigation of resveratrol and analogs as potential inducers of fetal hemoglobin. Blood Cells Mol. Dis. 2016, 58, 6–12. [Google Scholar] [CrossRef]
- Melo, T.R.F.; Kumkhaek, C.; Fernandes, G.F.D.S.; Lopes Pires, M.E.; Chelucci, R.C.; Barbieri, K.P.; Coelho, F.; Capote, T.S.O.; Lanaro, C.; Carlos, I.Z.; et al. Discovery of phenylsulfonylfuroxan derivatives as gamma globin inducers by histone acetylation. Eur. J. Med. Chem. 2018, 154, 341–353. [Google Scholar] [CrossRef] [PubMed]
- Iftikhar, F.; Ali, H.; Musharraf, S.G. Cinchona alkaloids as natural fetal hemoglobin inducing agents in human erythroleukemia cells. RSC Adv. 2019, 9, 17551–17559. [Google Scholar] [CrossRef]
- Nuamsee, K.; Chuprajob, T.; Pabuprapap, W.; Jintaridth, P.; Munkongdee, T.; Phannasil, P.; Vadolas, J.; Chaichompoo, P.; Suksamrarn, A.; Svasti, S. Trienone analogs of curcuminoids induce fetal hemoglobin synthesis via demethylation at Gγ-globin gene promoter. Sci. Rep. 2021, 11, 8552. [Google Scholar] [CrossRef]
- Ali, H.; Khan, F.; Ghulam Musharraf, S. Acyclovir induces fetal hemoglobin via downregulation of γ-globin repressors, BCL11A and SOX6 trans-acting factors. Biochem. Pharmacol. 2021, 190, 114612. [Google Scholar] [CrossRef]
- Gambari, R.; Fibach, E. Medicinal chemistry of fetal hemoglobin inducers for treatment of beta-thalassemia. Curr. Med. Chem. 2007, 14, 199–212. [Google Scholar] [CrossRef]
- Mischiati, C.; Sereni, A.; Lampronti, I.; Bianchi, N.; Borgatti, M.; Prus, E.; Fibach, E.; Gambari, R. Rapamycin-mediated induction of gamma-globin mRNA accumulation in human erythroid cells. Br. J. Haematol. 2004, 126, 612–621. [Google Scholar] [CrossRef] [PubMed]
- Zuccato, C.; Bianchi, N.; Borgatti, M.; Lampronti, I.; Massei, F.; Favre, C.; Gambari, R. Everolimus is a potent inducer of erythroid differentiation and gamma-globin gene expression in human erythroid cells. Acta Haematol. 2007, 117, 168–176. [Google Scholar] [CrossRef]
- Fibach, E.; Bianchi, N.; Borgatti, M.; Zuccato, C.; Finotti, A.; Lampronti, I.; Prus, E.; Mischiati, C.; Gambari, R. Effects of rapamycin on accumulation of alpha-, beta- and gamma-globin mRNAs in erythroid precursor cells from beta-thalassaemia patients. Eur. J. Haematol. 2006, 77, 437–441. [Google Scholar] [CrossRef] [PubMed]
- Pecoraro, A.; Troia, A.; Calzolari, R.; Scazzone, C.; Rigano, P.; Martorana, A.; Sacco, M.; Maggio, A.; Di Marzo, R. Efficacy of Rapamycin as Inducer of Hb F in Primary Erythroid Cultures from Sickle Cell Disease and β-Thalassemia Patients. Hemoglobin 2015, 39, 225–229. [Google Scholar] [CrossRef]
- Zhang, X.; Campreciós, G.; Rimmelé, P.; Liang, R.; Yalcin, S.; Mungamuri, S.K.; Barminko, J.; D’Escamard, V.; Baron, M.H.; Brugnara, C.; et al. FOXO3-mTOR metabolic cooperation in the regulation of erythroid cell maturation and homeostasis. Am. J. Hematol. 2014, 89, 954–963. [Google Scholar] [CrossRef] [PubMed]
- Khaibullina, A.; Almeida, L.E.; Wang, L.; Kamimura, S.; Wong, E.C.; Nouraie, M.; Maric, I.; Albani, S.; Finkel, J.; Quezado, Z.M. Rapamycin increases fetal hemoglobin and ameliorates the nociception phenotype in sickle cell mice. Blood Cells Mol. Dis. 2015, 55, 363–372. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Tran, J.; Wang, H.; Guo, C.; Harro, D.; Campbell, A.D.; Eitzman, D.T. mTOR Inhibition improves anaemia and reduces organ damage in a murine model of sickle cell disease. Br. J. Haematol. 2016, 174, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Lechauve, C.; Keith, J.; Khandros, E.; Fowler, S.; Mayberry, K.; Freiwan, A.; Thom, C.S.; Delbini, P.; Romero, E.B.; Zhang, J.; et al. The autophagy-activating kinase ULK1 mediates clearance of free α-globin in β-thalassemia. Sci. Transl. Med. 2019, 11, eaav4881. [Google Scholar] [CrossRef] [PubMed]
- Gamberini, M.R.; Prosdocimi, M.; Gambari, R. Sirolimus for Treatment of β-Thalassemia: From Pre-Clinical Studies to the Design of Clinical Trials. Health Educ. Public. Health 2021, 4, 425–435. [Google Scholar] [CrossRef]
- Zuccato, C.; Cosenza, L.C.; Zurlo, M.; Gasparello, J.; Papi, C.; D’Aversa, E.; Breveglieri, G.; Lampronti, I.; Finotti, A.; Borgatti, M.; et al. Expression of γ-globin genes in β-thalassemia patients treated with sirolimus: Results from a pilot clinical trial (Sirthalaclin). Ther. Adv. Hematol. 2022, 13, 20406207221100648. [Google Scholar] [CrossRef]
- Gaudre, N.; Cougoul, P.; Bartolucci, P.; Dörr, G.; Bura-Riviere, A.; Kamar, N.; Del Bello, A. Improved Fetal Hemoglobin with mTOR Inhibitor-Based Immunosuppression in a Kidney Transplant Recipient with Sickle Cell Disease. Am. J. Transplant. 2017, 17, 2212–2214. [Google Scholar] [CrossRef]
- Al-Khatti, A.A.; Alkhunaizi, A.M. Additive effect of sirolimus and hydroxycarbamide on fetal haemoglobin level in kidney transplant patients with sickle cell disease. Br. J. Haematol. 2019, 185, 959–961. [Google Scholar] [CrossRef] [PubMed]
- Zuccato, C.; Cosenza, L.C.; Zurlo, M.; Lampronti, I.; Borgatti, M.; Scapoli, C.; Gambari, R.; Finotti, A. Treatment of Erythroid Precursor Cells from β-Thalassemia Patients with Cinchona Alkaloids: Induction of Fetal Hemoglobin Production. Int. J. Mol. Sci. 2021, 22, 13433. [Google Scholar] [CrossRef]
- Cosenza, L.C.; Zuccato, C.; Zurlo, M.; Gambari, R.; Finotti, A. Co-Treatment of Erythroid Cells from β-Thalassemia Patients with CRISPR-Cas9-Based β039-Globin Gene Editing and Induction of Fetal Hemoglobin. Genes 2022, 13, 1727. [Google Scholar] [CrossRef]
- Ricchi, P.; Meloni, A.; Rigano, P.; Pistoia, L.; Spasiano, A.; Allò, M.; Messina, G.; Quarta, A.; Rosso, R.; Quota, A.; et al. The use of hydroxyurea in the real life of MIOT network: An observational study. Expert. Opin. Drug Saf. 2022, 21, 1433–1440. [Google Scholar] [CrossRef]
- Nag, A.; Radhakrishnan, V.S.; Kumar, J.; Bhave, S.; Mishra, D.K.; Nair, R.; Chandy, M. Thalidomide in Patients with Transfusion-Dependent E-Beta Thalassemia Refractory to Hydroxyurea: A Single-Center Experience. Indian J. Hematol. Blood Transfus. 2020, 36, 399–402. [Google Scholar] [CrossRef]
- Kosaryan, M.; Zafari, M.; Alipur, A.; Hedayatizadeh-Omran, A. The effect and side effect of hydroxyurea therapy on patients with β-thalassemia: A systematic review to December 2012. Hemoglobin 2014, 38, 262–271. [Google Scholar] [CrossRef]
- Konstantinou, E.; Pashalidis, I.; Kolnagou, A.; Kontoghiorghes, G.J. Interactions of hydroxycarbamide (hydroxyurea) with iron and copper: Implications on toxicity and therapeutic strategies. Hemoglobin 2011, 35, 237–246. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, N.; Finotti, A.; Ferracin, M.; Lampronti, I.; Zuccato, C.; Breveglieri, G.; Brognara, E.; Fabbri, E.; Borgatti, M.; Negrini, M.; et al. Increase of microRNA-210, decrease of raptor gene expression and alteration of mammalian target of rapamycin regulated proteins following mithramycin treatment of human erythroid cells. PLoS ONE 2015, 10, e0121567. [Google Scholar] [CrossRef]
- Finotti, A.; Bianchi, N.; Fabbri, E.; Borgatti, M.; Breveglieri, G.; Gasparello, J.; Gambari, R. Erythroid induction of K562 cells treated with mithramycin is associated with inhibition of raptor gene transcription and mammalian target of rapamycin complex 1 (mTORC1) functions. Pharmacol. Res. 2015, 91, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Prosdocimi, M.; Zuccato, C.; Cosenza, L.C.; Borgatti, M.; Lampronti, I.; Finotti, A.; Gambari, R. A Rational Approach to Drug Repositioning in β-thalassemia: Induction of Fetal Hemoglobin by Established Drugs. Wellcome Open Res. 2022, 7, 150. [Google Scholar] [CrossRef] [PubMed]
- Gasparello, J.; Fabbri, E.; Bianchi, N.; Breveglieri, G.; Zuccato, C.; Borgatti, M.; Gambari, R.; Finotti, A. BCL11A mRNA Targeting by miR-210: A Possible Network Regulating γ-Globin Gene Expression. Int. J. Mol. Sci. 2017, 18, 2530. [Google Scholar] [CrossRef]
- Cosenza, L.C.; Gasparello, J.; Romanini, N.; Zurlo, M.; Zuccato, C.; Gambari, R.; Finotti, A. Efficient CRISPR-Cas9-based genome editing of β-globin gene on erythroid cells from homozygous β039-thalassemia patients. Mol. Ther. Methods Clin. Dev. 2021, 21, 507–523. [Google Scholar] [CrossRef]
- Pule, G.D.; Mowla, S.; Novitzky, N.; Wiysonge, C.S.; Wonkam, A. A systematic review of known mechanisms of hydroxyurea-induced fetal hemoglobin for treatment of sickle cell disease. Expert. Rev. Hematol. 2015, 8, 669–679. [Google Scholar] [CrossRef]
- Bianchi, N.; Cosenza, L.C.; Lampronti, I.; Finotti, A.; Breveglieri, G.; Zuccato, C.; Fabbri, E.; Marzaro, G.; Chilin, A.; De Angelis, G.; et al. Structural and Functional Insights on an Uncharacterized Aγ-Globin-Gene Polymorphism Present in Four β0-Thalassemia Families with High Fetal Hemoglobin Levels. Mol. Diagn. Ther. 2016, 20, 161–173. [Google Scholar] [CrossRef] [PubMed]
- Breveglieri, G.; Bianchi, N.; Cosenza, L.C.; Gamberini, M.R.; Chiavilli, F.; Zuccato, C.; Montagner, G.; Borgatti, M.; Lampronti, I.; Finotti, A.; et al. An Aγ-globin G->A gene polymorphism associated with β039 thalassemia globin gene and high fetal hemoglobin production. BMC Med. Genet. 2017, 18, 93. [Google Scholar] [CrossRef] [PubMed]
- Prasing, W.; Odawara, T.; Traisathit, P.; Yamashiro, Y.; Hattori, Y.; Pornprasert, S. Analysis of theXmn1-Gγ polymorphism in β-thalassemia/hemoglobin E (HbE) and homozygous HbE patients with low and high levels of HbF. Int. J. Lab. Hematol. 2015, 37, e25–e28. [Google Scholar] [CrossRef] [PubMed]
- Zuccato, C.; Cosenza, L.C.; Zurlo, M.; Breveglieri, G.; Bianchi, N.; Lampronti, I.; Gasparello, J.; Scapoli, C.; Borgatti, M.; Finotti, A.; et al. The rs368698783 (G > A) Polymorphism Affecting LYAR Binding to the Aγ-Globin Gene Is Associated with High Fetal Hemoglobin (HbF) in β-Thalassemia Erythroid Precursor Cells Treated with HbF Inducers. Int. J. Mol. Sci. 2023, 24, 776. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gambari, R.; Zuccato, C.; Cosenza, L.C.; Zurlo, M.; Gasparello, J.; Finotti, A.; Gamberini, M.R.; Prosdocimi, M. The Long Scientific Journey of Sirolimus (Rapamycin): From the Soil of Easter Island (Rapa Nui) to Applied Research and Clinical Trials on β-Thalassemia and Other Hemoglobinopathies. Biology 2023, 12, 1202. https://doi.org/10.3390/biology12091202
Gambari R, Zuccato C, Cosenza LC, Zurlo M, Gasparello J, Finotti A, Gamberini MR, Prosdocimi M. The Long Scientific Journey of Sirolimus (Rapamycin): From the Soil of Easter Island (Rapa Nui) to Applied Research and Clinical Trials on β-Thalassemia and Other Hemoglobinopathies. Biology. 2023; 12(9):1202. https://doi.org/10.3390/biology12091202
Chicago/Turabian StyleGambari, Roberto, Cristina Zuccato, Lucia Carmela Cosenza, Matteo Zurlo, Jessica Gasparello, Alessia Finotti, Maria Rita Gamberini, and Marco Prosdocimi. 2023. "The Long Scientific Journey of Sirolimus (Rapamycin): From the Soil of Easter Island (Rapa Nui) to Applied Research and Clinical Trials on β-Thalassemia and Other Hemoglobinopathies" Biology 12, no. 9: 1202. https://doi.org/10.3390/biology12091202