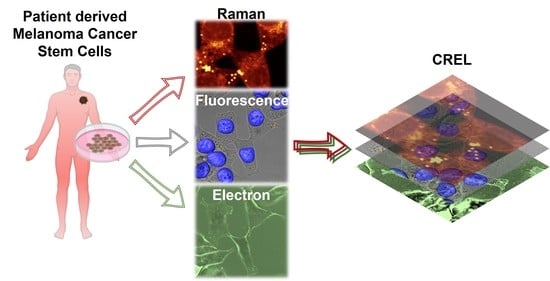
Correlative Raman–Electron–Light (CREL) Microscopy Analysis of Lipid Droplets in Melanoma Cancer Stem Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patterned Coverslips Preparation
2.2. Patient-Derived Melanoma Cancer Stem Cell Isolation and Characterization
2.3. Flow Cytometry
2.4. Raman Measurements
2.5. Raman Spectra Processing
2.6. Confocal Sample Preparation and Imaging
2.7. SEM Sample Preparation and Imaging
3. Results and Discussion
3.1. CREL Workflow
3.2. Lipid Droplet Analysis by Raman Spectroscopy
3.3. Lipid Droplet Analysis by Confocal Microscopy
3.4. Lipid Droplet Analysis by Scanning Electron Microscopy
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brinckerhoff, C.E. Cancer Stem Cells (CSCs) in melanoma: There’s smoke, but is there fire? J. Cell. Physiol. 2017, 232, 2674–2678. [Google Scholar] [CrossRef] [PubMed]
- Quintana, E.; Shackleton, M.; Sabel, M.S.; Fullen, D.R.; Johnson, T.M.; Morrison, S.J. Efficient tumour formation by single human melanoma cells. Nature 2008, 456, 593–598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, D.; Nguyen, T.K.; Leishear, K.; Finko, R.; Kulp, A.N.; Hotz, S.; Van Belle, P.A.; Xu, X.; Elder, D.E.; Herlyn, M. A tumorigenic subpopulation with stem cell properties in melanomas. Cancer Res. 2005, 65, 9328–9337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marzagalli, M.; Raimondi, M.; Fontana, F.; Marelli, M.M.; Moretti, R.M.; Limonta, P. Cellular and molecular biology of cancer stem cells in melanoma: Possible therapeutic implications. Semin. Cancer Biol. 2019, 59, 221–235. [Google Scholar] [CrossRef]
- Giampietri, C.; Petrungaro, S.; Cordella, M.; Tabolacci, C.; Tomaipitinca, L.; Facchiano, A.; Eramo, A.; Filippini, A.; Facchiano, F.; Ziparo, E. Lipid storage and autophagy in melanoma cancer cells. Int. J. Mol. Sci. 2017, 18, 1271. [Google Scholar] [CrossRef] [Green Version]
- Tirinato, L.; Liberale, C.; Di Franco, S.; Candeloro, P.; Benfante, A.; La Rocca, R.; Potze, L.; Marotta, R.; Ruffilli, R.; Rajamanickam, V.P.; et al. Lipid droplets: A new player in colorec-tal cancer stem cells unveiled by spectroscopic imaging. Stem Cells 2015, 33, 35–44. [Google Scholar] [CrossRef] [Green Version]
- Tirinato, L.; Pagliari, F.; Limongi, T.; Marini, M.; Falqui, A.; Seco, J.; Candeloro, P.; Liberale, C.; Di Fabrizio, E. An overview of lipid droplets in cancer and cancer stem cells. Stem Cells Int. 2017, 2017, 1656053. [Google Scholar] [CrossRef] [Green Version]
- Tirinato, L.; Pagliari, F.; Di Franco, S.; Sogne, E.; Marafioti, M.G.; Jansen, J.; Falqui, A.; Todaro, M.; Candeloro, P.; Liberale, C.; et al. ROS and Lipid Droplet accumulation induced by high glucose exposure in healthy colon and Colorectal Cancer Stem Cells. Genes Dis. 2020, 7, 620–635. [Google Scholar] [CrossRef]
- Tirinato, L.; Marafioti, M.G.; Pagliari, F.; Jansen, J.; Aversa, I.; Hanley, R.; Nisticò, C.; Garcia-Calderón, D.; Genard, G.; Guerreiro, J.F.; et al. Lipid droplets and ferritin heavy chain: A devilish liaison in human cancer cell radioresistance. Elife 2021, 10, e72943. [Google Scholar] [CrossRef]
- Welte, M. Expanding roles for lipid droplets. Curr. Biol. 2015, 25, R470–R481. [Google Scholar] [CrossRef]
- Caplan, J.; Niethammer, M.; Taylor, R.M., II; Czymmek, K.J. The power of correla-tive microscopy: Multi-modal, multi-scale, multi-dimensional. Curr. Opin. Struct. Biol. 2011, 21, 686–693. [Google Scholar] [CrossRef] [Green Version]
- Van Rijnsoever, C.; Oorschot, V.; Klumperman, J. Correlative light-electron mi-croscopy (CLEM) combining live-cell imaging and immunolabeling of ultrathin cryo-sections. Nat. Methods 2008, 5, 973–980. [Google Scholar] [CrossRef]
- Mironov, A.A.; Beznoussenko, G.V. Correlative microscopy: A potent tool for the study of rare or unique cellular and tissue events. J. Microsc. 2009, 235, 308–321. [Google Scholar] [CrossRef]
- Schmidt, U.; Ayasse, P.; Hollricher, O. RISE Microscopy: Correlative R aman and SEM Imaging. In Proceedings of the European Microscopy Congress 2016, Lyon, France, 28 August–2 September 2016; Wiley Online Library: Weinheim, Germany, 2016; pp. 1023–1024. [Google Scholar]
- Timmermans, F.J.; Liszka, B.; Lenferink, A.T.; van Wolferen, H.A.; Otto, C. Integration of correlative Raman microscopy in a dualbeam FIB SEM. J. Raman. Spectrosc. 2016, 47, 956–962. [Google Scholar] [CrossRef]
- Enciso-Martinez, A.; Timmermans, F.J.; Nanou, A.; Terstappen, L.; Otto, C. SEM–Raman image cytometry of cells. Analyst 2018, 143, 4495–4502. [Google Scholar] [CrossRef]
- Beekman, P.; Enciso-Martinez, A.; Rho, H.S.; Pujari, S.P.; Lenferink, A.; Zuilhof, H.; Terstappen, L.W.; Otto, C.; Le Gac, S. Immuno-capture of extracellular vesicles for individual multi-modal characterization using AFM, SEM and Raman spectroscopy. Lab A Chip 2019, 19, 2526–2536. [Google Scholar] [CrossRef] [Green Version]
- Benedetti, L.; Sogne, E.; Rodighiero, S.; Marchesi, D.; Milani, P.; Francolini, M. Customized patterned substrates for highly versatile correlative light-scanning electron microscopy. Sci. Rep. 2014, 4, 7033. [Google Scholar] [CrossRef] [Green Version]
- Todaro, M.; Alea, M.P.; Di Stefano, A.B.; Cammareri, P.; Vermeulen, L.; Iovino, F.; Tripodo, C.; Russo, A.; Gulotta, G.; Medema, J.P.; et al. Colon cancer stem cells dictate tumor growth and resist cell death by production of interleukin-4. Cell Stem Cell 2007, 1, 389–402. [Google Scholar] [CrossRef] [Green Version]
- Candeloro, P.; Grande, E.; Raimondo, R.; Di Mascolo, D.; Gentile, F.; Coluccio, M.L.; Perozziello, G.; Malara, N.; Francardi, M.; Di Fabrizio, E. Raman database of amino acids solutions: A critical study of Extended Multiplicative Signal Correction. Analyst 2013, 138, 7331–7340. [Google Scholar] [CrossRef]
- Spandl, J.; White, D.J.; Peychl, J.; Thiele, C. Live cell multicolor imaging of lipid droplets with a new dye, LD540. Traffic 2009, 10, 1579–1584. [Google Scholar] [CrossRef]
- Krafft, C.; Knetschke, T.; Funk, R.H.; Salzer, R. Identification of organelles and vesicles in single cells by Raman microspectroscopic mapping. Vib. Spectrosc. 2005, 38, 85–93. [Google Scholar] [CrossRef]
- Read, D.S.; Woodcock, D.J.; Strachan, N.J.; Forbes, K.J.; Colles, F.M.; Maiden, M.C.; Clifton-Hadley, F.; Ridley, A.; Vidal, A.; Rodgers, J.; et al. Evidence for phenotypic plasticity among multihost Campylobacter jejuni and C. coli lineages, obtained using ribosomal multilocus sequence typing and Raman spectroscopy. Appl. Environ. Microbiol. 2013, 79, 965–973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johannessen, C.; White, P.C.; Abdali, S. Resonance Raman optical activity and surface enhanced resonance Raman optical activity analysis of cytochrome c. J. Phys. Chem. A 2007, 111, 7771–7776. [Google Scholar] [CrossRef]
- Sinibaldi, F.; Mei, G.; Polticelli, F.; Piro, M.C.; Howes, B.D.; Smulevich, G.; Santucci, R.; Ascoli, F.; Fiorucci, L. ATP specifically drives refolding of non-native conformations of cytochrome c. Protein Sci. 2005, 14, 1049–1058. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Czamara, K.; Majzner, K.; Pacia, M.Z.; Kochan, K.; Kaczor, A.; Baranska, M. Raman spectroscopy of lipids: A review. J. Raman Spectrosc. 2014, 46, 4–20. [Google Scholar] [CrossRef]
- Wu, H.; Volponi, J.V.; Oliver, A.E.; Parikh, A.N.; Simmons, B.A.; Singh, S. In vivo lipidomics using single-cell Raman spectroscopy. Proc. Natl. Acad. Sci. USA 2011, 108, 3809–3814. [Google Scholar] [CrossRef] [Green Version]
- Lippert, J.L.; Peticolas, W.L. Laser Raman investigation of the effect of cholesterol on conformational changes in dipalmitoyl lecithin multilayers. Proc. Natl. Acad. Sci. USA 1971, 68, 1572–1576. [Google Scholar] [CrossRef] [Green Version]
- Herms, A.; Bosch, M.; Reddy, B.J.; Schieber, N.L.; Fajardo, A.; Rupérez, C.; Fernán-dez-Vidal, A.; Ferguson, C.; Rentero, C.; Tebar, F.; et al. AMPK activation promotes lipid droplet dispersion on detyrosinated microtubules to increase mitochondrial fatty acid oxidation. Nat. Commun. 2015, 6, 7176. [Google Scholar] [CrossRef] [Green Version]
- Bradley, J.; Pope, I.; Masia, F.; Sanusi, R.; Langbein, W.; Swann, K.; Borri, P. Quantitative imaging of lipids in live mouse oocytes and early embryos using CARS microscopy. Development 2016, 143, 2238–2247. [Google Scholar] [CrossRef] [Green Version]
- Katsen-Globa, A.; Puetz, N.; Gepp, M.M.; Neubauer, J.C.; Zimmermann, H. Study of SEM preparation artefacts with correlative microscopy: Cell shrinkage of adherent cells by HMDS-drying. Scanning 2016, 38, 625–633. [Google Scholar] [CrossRef]
- Zhang, Y.; Huang, T.; Jorgens, D.M.; Nickerson, A.; Lin, L.-J.; Pelz, J.; Gray, J.W.; López, C.S.; Nan, X. Quantitating morphological changes in biological samples during scanning electron microscopy sample preparation with correlative super-resolution microscopy. PLoS ONE 2017, 12, e0176839. [Google Scholar] [CrossRef] [Green Version]
- Cheng, J.; Xie, X.S. Vibrational spectroscopic imaging of living systems: An emerging platform for biology and medicine. Science 2015, 350, aaa8870. [Google Scholar] [CrossRef]
- Bernhardt, M.; Nicolas, J.D.; Osterhoff, M.; Mittelstädt, H.; Reuss, M.; Harke, B.; Wittmeier, A.; Sprung, M.; Köster, S.; Salditt, T. Correlative microscopy approach for biology using X-ray holography, X-ray scanning diffraction and STED microscopy. Nat. Commun. 2018, 9, 3641. [Google Scholar] [CrossRef]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pagliari, F.; Sogne, E.; Panella, D.; Perozziello, G.; Liberale, C.; Das, G.; Turdo, A.; Di Franco, S.; Seco, J.; Falqui, A.; et al. Correlative Raman–Electron–Light (CREL) Microscopy Analysis of Lipid Droplets in Melanoma Cancer Stem Cells. Biosensors 2022, 12, 1102. https://doi.org/10.3390/bios12121102
Pagliari F, Sogne E, Panella D, Perozziello G, Liberale C, Das G, Turdo A, Di Franco S, Seco J, Falqui A, et al. Correlative Raman–Electron–Light (CREL) Microscopy Analysis of Lipid Droplets in Melanoma Cancer Stem Cells. Biosensors. 2022; 12(12):1102. https://doi.org/10.3390/bios12121102
Chicago/Turabian StylePagliari, Francesca, Elisa Sogne, Davide Panella, Gerardo Perozziello, Carlo Liberale, Gobind Das, Alice Turdo, Simone Di Franco, Joao Seco, Andrea Falqui, and et al. 2022. "Correlative Raman–Electron–Light (CREL) Microscopy Analysis of Lipid Droplets in Melanoma Cancer Stem Cells" Biosensors 12, no. 12: 1102. https://doi.org/10.3390/bios12121102