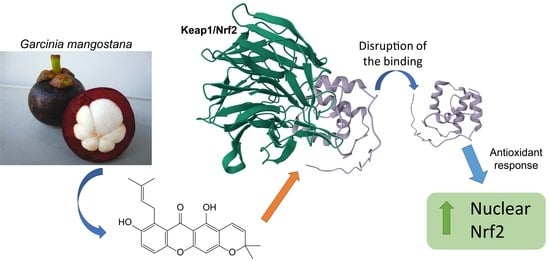
Demethylcalabaxanthone from Garcinia mangostana Exerts Antioxidant Effects through the Activation of the Nrf2 Pathway as Assessed via Molecular Docking and Biological Evaluation
Abstract
:1. Introduction
2. Materials and Methods
2.1. General Experimental Procedures
2.2. Plant Material
2.3. Extraction and Isolation
2.4. Molecular Docking
2.5. In Vitro Experiments
2.5.1. Cell Cultures and Treatment
2.5.2. Cytotoxicity Test
2.5.3. Cytoprotection against Oxidative Stress Induced by H2O2
2.5.4. LDH Liberation Test
2.5.5. Determination of Reduced Glutathione Level (GSH)
2.5.6. Quantitative Determination of Protein Content using the BCA Method
2.5.7. Protein Extraction
2.5.8. Protein Dosage Using the Bradford Method
2.5.9. Polyacrylamide Gel Electrophoresis (SDS-PAGE)
2.5.10. Western Blotting
2.5.11. Statistical Analysis
3. Results and Discussion
3.1. Extraction and Isolation
3.2. Molecular Docking
- P1: Arg415, Ile461, Gly462, Phe478, Arg483, and Ser508;
- P2: Ser363, Arg380, Asn382, and Asn414;
- P3: Gly509, Ser555, Ala556, Gly571, Ser602, and Gly603;
- P4: Tyr525, Gln530, and Tyr572;
- P5: Tyr334 and Phe577.
3.3. In Vitro Experiments
3.3.1. Cytotoxicity Test
3.3.2. Cytoprotection against Oxidative Stress Induced by H2O2
3.3.3. Determination of Reduced Glutathione Level (GSH)
3.3.4. LDH Liberation Test
3.3.5. Protein Analysis (Western Blotting)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ma, Q. Transcriptional responses to oxidative stress: Pathological and toxicological implications. Pharmacol. Ther. 2010, 125, 376–393. [Google Scholar] [CrossRef] [PubMed]
- Choi, B.-H.; Kang, K.-S.; Kwak, M.-K. Effect of redox modulating NRF2 activators on chronic kidney disease. Mol. Cells 2014, 19, 12727–12759. [Google Scholar] [CrossRef] [PubMed]
- Circu, M.L.; Aw, T.Y. Reactive oxygen species, cellular redox systems, and apoptosis. Free Radic. Biol. Med. 2010, 48, 749–762. [Google Scholar] [CrossRef] [PubMed]
- Valko, M.; Leibfritz, D.; Moncol, J.; Cronin, M.T.D.; Mazur, M.; Telser, J. Free radicals and antioxidants in normal physiological functions and human disease. Int. J. Biochem. Cell Biol. 2007, 39, 44–84. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q. Role of Nrf2 in oxidative stress and toxicity. Annu. Rev. Pharmacol. Toxicol. 2013, 53, 401–426. [Google Scholar] [CrossRef]
- Bello, M.; Morales-González, J.A. Molecular recognition between potential natural inhibitors of the Keap1-Nrf2 complex. Int. J. Biol. Macromol. 2017, 105, 981–992. [Google Scholar] [CrossRef] [PubMed]
- Sihvola, V.; Levonen, A.-L. Keap1 as the redox sensor of the antioxidant response. Arch. Biochem. Biophys. 2017, 617, 94–100. [Google Scholar] [CrossRef]
- Kwak, M.-K.; Itoh, K.; Yamamoto, M.; Sutter, T.R.; Kensler, T.W. Role of transcription factor Nrf2 in the induction of hepatic phase 2 and antioxidative enzymes in vivo by the cancer chemoprotective agent, 3H-1, 2-dithiole-3-thione. Mol. Med. Rep. 2001, 7, 135–145. [Google Scholar] [CrossRef]
- Zhang, D.D. Mechanistic studies of the Nrf2-Keap1 signaling pathway. Drug Metab. Rev. 2006, 38, 769–789. [Google Scholar] [CrossRef]
- Zhuang, C.; Narayanapillai, S.; Zhang, W.; Sham, Y.Y.; Xing, C. Rapid identification of Keap1–Nrf2 small-molecule inhibitors through structure-based virtual screening and hit-based substructure search. J. Med. Chem. 2014, 57, 1121–1126. [Google Scholar] [CrossRef]
- Li, J.; Deng, S.-h.; Li, J.; Li, L.; Zhang, F.; Zou, Y.; Wu, D.-M.; Xu, Y. Obacunone alleviates ferroptosis during lipopolysaccharide-induced acute lung injury by upregulating Nrf2-dependent antioxidant responses. Cell Mol. Biol. Lett. 2022, 27, 29. [Google Scholar] [CrossRef]
- Dang, R.; Wang, M.; Li, X.; Wang, H.; Liu, L.; Wu, Q.; Zhao, J.; Ji, P.; Zhong, L.; Licinio, J.; et al. Edaravone ameliorates depressive and anxiety-like behaviors via Sirt1/Nrf2/HO-1/Gpx4 pathway. J. Neuroinflamm. 2022, 19, 41. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, W.-J.; Zheng, X.-R.; Liu, Q.-L.; Du, Q.; Lai, Y.-J.; Liu, S.-Q. Eriodictyol ameliorates cognitive dysfunction in APP/PS1 mice by inhibiting ferroptosis via vitamin D receptor-mediated Nrf2 activation. Mol. Med. 2022, 28, 11. [Google Scholar] [CrossRef] [PubMed]
- Abate, M.; Pagano, C.; Masullo, M.; Citro, M.; Pisanti, S.; Piacente, S.; Bifulco, M. Mangostanin, a xanthone derived from Garcinia mangostana fruit, exerts protective and reparative effects on oxidative damage in human keratinocytes. Pharmaceuticals 2022, 15, 84. [Google Scholar] [CrossRef] [PubMed]
- Gallorini, M.; Carradori, S.; Resende, D.; Saso, L.; Ricci, A.; Palmeira, A.; Cataldi, A.; Pinto, M.; Sousa, E. Natural and synthetic xanthone derivatives counteract oxidative stress via Nrf2 modulation in inflamed human macrophages. Int. J. Mol. Sci. 2022, 23, 13319. [Google Scholar] [CrossRef] [PubMed]
- Ovalle-Magallanes, B.; Eugenio-Pérez, D.; Pedraza-Chaverri, J. Medicinal properties of mangosteen (Garcinia mangostana L.): A comprehensive update. Food Chem. Toxicol. 2017, 109, 102–122. [Google Scholar] [CrossRef]
- Nauman, M.C.; Johnson, J.J. The purple mangosteen (Garcinia mangostana): Defining the anticancer potential of selected xanthones. Pharmacol. Res. 2022, 175, 106032. [Google Scholar] [CrossRef] [PubMed]
- Yazarlu, O.; Iranshahi, M.; Kashani, H.R.K.; Reshadat, S.; Habtemariam, S.; Iranshahy, M.; Hasanpour, M. Perspective on the application of medicinal plants and natural products in wound healing: A mechanistic review. Pharmacol. Res. 2021, 174, 105841. [Google Scholar] [CrossRef]
- Ma, B.; Lucas, B.; Capacci, A.; Lin, E.Y.-S.; Jones, J.H.; Dechantsreiter, M.; Enyedy, I.; Marcotte, D.; Xiao, G.; Li, B.; et al. Design, synthesis and identification of novel, orally bioavailable non-covalent Nrf2 activators. Bioorg. Med. Chem. Lett. 2020, 30, 126852. [Google Scholar] [CrossRef]
- Schrödinger Release 2020-1: Protein Preparation Wizard; Schrödinger LLC: New York, NY, USA, 2020.
- Schrödinger Release 2020-1: Maestro; Schrödinger LLC: New York, NY, USA, 2020.
- Schrödinger Release 2020-1: LigPrep; Schrödinger LLC: New York, NY, USA, 2020.
- Schrödinger Release 2020-1: Glide; Schrödinger LLC: New York, NY, USA, 2020.
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef]
- Halgren, T.A.; Murphy, R.B.; Friesner, R.A.; Beard, H.S.; Frye, L.L.; Pollard, W.T.; Banks, J.L. Glide: A new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J. Med. Chem. 2004, 47, 1750–1759. [Google Scholar] [CrossRef] [PubMed]
- Bermejo-Bescós, P.; Piñero-Estrada, E.; del Fresno, Á.M.V. Neuroprotection by Spirulina platensis protean extract and phycocyanin against iron-induced toxicity in SH-SY5Y neuroblastoma cells. Toxicol. In Vitro 2008, 22, 1496–1502. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Su, T.; Qiu, X.; Mao, P.; Xu, Y.; Hu, Z.; Zhang, Y.; Zheng, X.; Xie, P.; Liu, Q. Protective effect of alpha-mangostin against oxidative stress induced-retinal cell death. Sci. Rep. 2016, 6, 21018. [Google Scholar] [CrossRef] [PubMed]
- Masullo, M.; Cerulli, A.; Cannavacciuolo, C.; Kılınç, H.; Pizza, C.; Piacente, S. Garcinia mangostana L. fruits and derived food supplements: Identification and quantitative determination of bioactive xanthones by NMR analysis. J. Pharm. Biomed. Anal. 2022, 218, 114835. [Google Scholar] [CrossRef]
- Lo, S.C.; Li, X.; Henzl, M.T.; Beamer, L.J.; Hannink, M. Structure of the Keap1: Nrf2 interface provides mechanistic insight into Nrf2 signaling. EMBO J. 2006, 25, 3605–3617. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.-R.; Hu, L.-S.; Li, G.-Y. SH-SY5Y human neuroblastoma cell line: In vitrocell model of dopaminergic neurons in Parkinson’s disease. Chin. Med. J. 2010, 123, 1086–1092. [Google Scholar] [CrossRef]
- Houck, A.L.; Hernández, F.; Ávila, J. A simple model to study tau pathology. J. Exp. Neurosci. 2016, 10, 31–38. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Ngo, V.; Duennwald, M.L. Nrf2 and oxidative stress: A general overview of mechanisms and implications in human disease. Antioxidants 2022, 11, 2345. [Google Scholar] [CrossRef]
- Smith, P.K.; Krohn, R.I.; Hermanson, G.T.; Mallia, A.K.; Gartner, F.H.; Provenzano, M.D.; Fujimoto, E.K.; Goeke, N.M.; Olson, B.J.; Klenk, D.C. Measurement of protein using bicinchoninic acid. Anal. Biochem. 1985, 150, 76–85. [Google Scholar] [CrossRef] [PubMed]
- Sun, A.Y.; Chen, Y.-M.; James-Kracke, M.; Wixom, P.; Cheng, Y. Ethanol-induced cell death by lipid peroxidation in PC12 cells. Neurochem. Res. 1997, 22, 1187–1192. [Google Scholar] [CrossRef] [PubMed]
- Brackhan, M.; Arribas-Blazquez, M.; Lastres-Becker, I. Aging, Nrf2, and TAU: A perfect match for neurodegeneration? Antioxidants 2023, 12, 1564. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, G.A.; Ibrahim, S.R.M.; Hareeri, R.H.; Binmahfouz, L.S.; Bagher, A.M.; Abdallah, H.M.; Elsaed, W.M.; El-Agamy, D.S. Garcinone E mitigates oxidative inflammatory response and protects against experimental autoimmune hepatitis via modulation of Nrf2/HO-1, NF-kappaB and TNF-alpha/JNK axis. Nutrients 2023, 15, 16. [Google Scholar] [CrossRef]
- John, O.D.; Mushunje, A.T.; Surugau, N.; Guad, R.M. The metabolic and molecular mechanisms of α-mangostin in cardiometabolic disorders (Review). Int. J. Mol. Med. 2022, 50, 120. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wang, S.; Ouyang, Y.; Tu, Y.; Liu, A.; Tian, Y.; He, M.; Pi, R. Garcinone D, a natural xanthone promotes C17.2 neural stem cell proliferation: Possible involvement of STAT3/Cyclin D1 pathway and Nrf2/HO-1 pathway. Neurosci. Lett. 2016, 626, 6–12. [Google Scholar] [CrossRef] [PubMed]
- Khayat, M.T.; Mohammad, K.A.; Mohamed, G.A.; El-Agamy, D.S.; Elsaed, W.M.; Ibrahim, S.R.M. γ-Mangostin abrogates AINT-induced cholestatic liver injury: Impact on Nrf2/NF-κB/NLRP3/Caspase-1/IL-1β/GSDMD signalling. Life Sci. 2023, 322, 121663. [Google Scholar] [CrossRef] [PubMed]







| Compounds | H-Bond | π-Cation | π-π Stacking | Hydrophobic |
|---|---|---|---|---|
| 1 | Ser363 and Asn414 | Arg415 (2) | Tyr334 and Tyr525 | Gly364, Arg380, Gly462, Arg483, Ser508, Gly509, Gln530, Ser555, Ala556, Tyr572, Phe577, Ser602, and Gly603 |
| 2 | Ser363, Asn414, and Ser508 | Arg415 (2) | / | Tyr334, Ser338, Gly364, Arg380, Asn382, Gly462, Gly509, Tyr525, Gln530, Ser555, Ala556, Tyr572, Phe577, and Ser602 |
| 3 | Ser508 | Arg415 (2) | Tyr525 | Tyr334, Ser363, Gly364, Arg380, Asn382, Asn414, Ile416, Gly462, Arg483, Gly509, Ala510, Gln530, Ser555, Ala556, Leu557, Tyr572, Phe577, Ser602, and Gly603 |
| 4 | Ser363 and Ser602 | Arg415 (2) | Tyr334 and Tyr572 | Gly364, Arg380, Asn382, Asn414, Gly462, Gly509, Ala556, Phe577, and Gly603 |
| 5 | Ser363 and Ser508 | / | Tyr525 | Tyr334, Gly364, Arg380, Asn382, Asn414, Arg415, Ile461, Gly462, Phe478, Arg483, Gly509, Gln530, Ser555, Ala556, Tyr572, Phe577, Ser602, and Gly603 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Vita, S.; Masullo, M.; Grambone, S.; Bescós, P.B.; Piacente, S.; Bifulco, G. Demethylcalabaxanthone from Garcinia mangostana Exerts Antioxidant Effects through the Activation of the Nrf2 Pathway as Assessed via Molecular Docking and Biological Evaluation. Antioxidants 2023, 12, 1980. https://doi.org/10.3390/antiox12111980
De Vita S, Masullo M, Grambone S, Bescós PB, Piacente S, Bifulco G. Demethylcalabaxanthone from Garcinia mangostana Exerts Antioxidant Effects through the Activation of the Nrf2 Pathway as Assessed via Molecular Docking and Biological Evaluation. Antioxidants. 2023; 12(11):1980. https://doi.org/10.3390/antiox12111980
Chicago/Turabian StyleDe Vita, Simona, Milena Masullo, Sabrina Grambone, Paloma Bermejo Bescós, Sonia Piacente, and Giuseppe Bifulco. 2023. "Demethylcalabaxanthone from Garcinia mangostana Exerts Antioxidant Effects through the Activation of the Nrf2 Pathway as Assessed via Molecular Docking and Biological Evaluation" Antioxidants 12, no. 11: 1980. https://doi.org/10.3390/antiox12111980