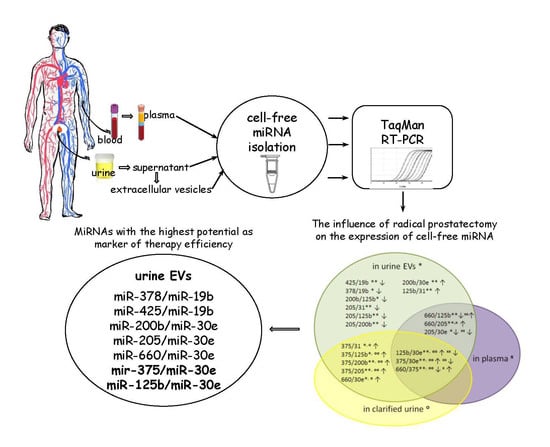
The Influence of Radical Prostatectomy on the Expression of Cell-Free MiRNA
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Isolation of Urine EVs by Ultracentrifugation
2.3. Isolation of miRNAs by the Gu/OcA Protocol
2.4. RNA Precipitation
2.5. Reverse Transcription and Quantitative RT–PCR
2.6. Statistical Analysis
3. Results
3.1. The Influence of the RP on the Expression of Cell-Free miRNA
3.2. Comparative Analysis of the miRNA Expression in the Biofluids of PCa Patients after RP and in That of Healthy Donors
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Tourinho-Barbosa, R.; Srougi, V.; Nunes-Silva, I.; Baghdadi, M.; Rembeyo, G.; Eiffel, S.S.; Barret, E.; Rozet, F.; Galiano, M.; Cathelineau, X.; et al. Biochemical recurrence after radical prostatectomy: What does it mean? Int. Braz. J. Urol. 2018, 44, 14–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Armand-Labit, V.; Pradines, A. Circulating cell-free microRNAs as clinical cancer biomarkers. Biomol. Concepts 2017, 24, 61–81. [Google Scholar] [CrossRef] [PubMed]
- Bryzgunova, O.E.; Laktionov, P.P. Extracellular nucleic acids in urine: Sources, structure, diagnostic potential. Acta Nat. 2015, 7, 48–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barron, N.; Keenan, J.; Gammell, P.; Martinez, V.G.; Freeman, A.; Masters, J.R.; Clynes, M. Biochemical relapse following radical prostatectomy and miR-200a levels in prostate cancer. Prostate 2012, 72, 1193–1199. [Google Scholar] [CrossRef] [PubMed]
- Casanova-Salas, I.; Rubio-Briones, J.; Calatrava, A.; Mancarella, C.; Masiá, E.; Casanova, J.; Fernández-Serra, A.; Rubio, L.; Ramírez-Backhaus, M.; Armiñán, A.; et al. Identification of miR-187 and miR-182 as Biomarkers of Early Diagnosis and Prognosis in Patients with Prostate Cancer Treated with Radical Prostatectomy. J. Urol. 2014, 192, 252–259. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Peskoe, S.B.; Ribas, J.; Rafiqi, F.; Kudrolli, T.; Meeker, A.K.; De Marzo, A.M.; Platz, E.A.; Lupold, S.E. Investigation of miR-21, miR-141, and miR-221 expression levels in prostate adenocarcinoma for associated risk of recurrence after radical prostatectomy: miR-221 and Prostate Cancer Recurrence. Prostate 2014, 74, 1655–1662. [Google Scholar] [CrossRef] [Green Version]
- Mahn, R.; Heukamp, L.C.; Rogenhofer, S.; von Ruecker, A.; Müller, S.C.; Ellinger, J. Circulating microRNAs (miRNA) in Serum of Patients With Prostate Cancer. Urology 2011, 77, 1265.e9–1265.e16. [Google Scholar] [CrossRef]
- Egidi, M.G.; Cochetti, G.; Serva, M.R.; Guelfi, G.; Zampini, D.; Mechelli, L.; Mearini, E. Circulating microRNAs and Kallikreins before and after Radical Prostatectomy: Are They Really Prostate Cancer Markers? BioMed Res. Int. 2013, 2013, 1–11. [Google Scholar] [CrossRef]
- Zedan, A.H.; Hansen, T.F.; Assenholt, J.; Madsen, J.S.; Osther, P.J.S. Circulating miRNAs in localized/locally advanced prostate cancer patients after radical prostatectomy and radiotherapy. Prostate 2018. [Google Scholar] [CrossRef] [Green Version]
- Kossinova, O.A.; Gopanenko, A.V.; Tamkovich, S.N.; Krasheninina, O.A.; Tupikin, A.E.; Kiseleva, E.; Yanshina, D.D.; Malygin, A.A.; Ven’yaminova, A.G.; Kabilov, M.R.; et al. Cytosolic YB-1 and NSUN2 are the only proteins recognizing specific motifs present in mRNAs enriched in exosomes. Biochim. Biophys. Acta Proteins Proteom. 2017, 1865, 664–673. [Google Scholar] [CrossRef]
- Bryzgunova, O.E.; Zaripov, M.M.; Skvortsova, T.E.; Lekchnov, E.A.; Grigor’eva, A.E.; Zaporozhchenko, I.A.; Morozkin, E.S.; Ryabchikova, E.I.; Yurchenko, Y.B.; Voitsitskiy, V.E.; et al. Comparative Study of Extracellular Vesicles from the Urine of Healthy Individuals and Prostate Cancer Patients. PLoS ONE 2016, 11, e0157566. [Google Scholar] [CrossRef] [PubMed]
- Fredsøe, J.; Rasmussen, A.K.I.; Thomsen, A.R.; Mouritzen, P.; Høyer, S.; Borre, M.; Ørntoft, T.F.; Sørensen, K.D. Diagnostic and Prognostic MicroRNA Biomarkers for Prostate Cancer in Cell-free Urine. Eur. Urol. Focus 2018, 4, 825–833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bryzgunova, O.E.; Zaporozhchenko, I.A.; Lekchnov, E.A.; Amelina, E.V.; Konoshenko, M.Y.; Yarmoschuk, S.V.; Pashkovskaya, O.A.; Zheravin, A.A.; Pak, S.V.; Rykova, E.Y.; et al. Data analysis algorithm for the development of extracellular miRNA-based diagnostic systems for prostate cancer. PLoS ONE 2019, 14, e0215003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lekchnov, E.A.; Amelina, E.V.; Bryzgunova, O.E.; Zaporozhchenko, I.A.; Konoshenko, M.Y.; Yarmoschuk, S.V.; Murashov, I.S.; Pashkovskaya, O.A.; Gorizkii, A.M.; Zheravin, A.A.; et al. Searching for the Novel Specific Predictors of Prostate Cancer in Urine: The Analysis of 84 miRNA Expression. Int. J. Mol. Sci. 2018, 19, 4088. [Google Scholar] [CrossRef] [Green Version]
- Konoshenko, M.Y.; Lekchnov, E.A.; Bryzgunova, O.E.; Zaporozhchenko, I.A.; Yarmoschuk, S.V.; Pashkovskaya, O.A.; Pak, S.V.; Laktionov, P.P. The Panel of 12 Cell-Free MicroRNAs as Potential Biomarkers in Prostate Neoplasms. Diagnostics 2020, 10, 38. [Google Scholar] [CrossRef] [Green Version]
- Lekchnov, E.A.; Zaporozhchenko, I.A.; Morozkin, E.S.; Bryzgunova, O.E.; Vlassov, V.V.; Laktionov, P.P. Protocol for miRNA isolation from biofluids. Anal. Biochem. 2016, 499, 78–84. [Google Scholar] [CrossRef]
- Boeri, M.; Verri, C.; Conte, D.; Roz, L.; Modena, P.; Facchinetti, F.; Calabrò, E.; Croce, C.M.; Pastorino, U.; Sozzi, G. MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc. Natl. Acad. Sci. USA 2011, 108, 3713–3718. [Google Scholar] [CrossRef] [Green Version]
- Landoni, E.; Miceli, R.; Callari, M.; Tiberio, P.; Appierto, V.; Angeloni, V.; Mariani, L.; Daidone, M.G. Proposal of supervised data analysis strategy of plasma miRNAs from hybridisation array data with an application to assess hemolysis-related deregulation. BMC Bioinform. 2015, 16, 388. [Google Scholar] [CrossRef] [Green Version]
- Kelly, B.; Miller, N.; Sweeney, K.; Durkan, G.; Rogers, E.; Walsh, K.; Kerin, M. A Circulating MicroRNA Signature as a Biomarker for Prostate Cancer in a High Risk Group. JCM 2015, 4, 1369–1379. [Google Scholar] [CrossRef] [Green Version]
- McDonald, A.C.; Raman, J.D.; Shen, J.; Liao, J.; Pandya, B.; Vira, M.A. Circulating microRNAs in plasma before and after radical prostatectomy. Urol. Oncol. Semin. Ori. 2019, 37, 814.e1–814.e7. [Google Scholar] [CrossRef]
- Al-Qatati, A.; Akrong, C.; Stevic, I.; Pantel, K.; Awe, J.; Saranchuk, J.; Drachenberg, D.; Mai, S.; Schwarzenbach, H. Plasma microRNA signature is associated with risk stratification in prostate cancer patients. Int. J. Cancer 2017, 141, 1231–1239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.S.; Dutta, A. MicroRNAs in Cancer. Annu. Rev. Pathol. Mech. Dis. 2009, 4, 199–227. [Google Scholar] [CrossRef] [PubMed]
- Bryzgunova, O.E.; Konoshenko, M.Y.; Laktionov, P.P. MicroRNA-guided gene expression in prostate cancer: Literature and database overview. J. Gene Med. 2018, 20, e3016. [Google Scholar] [CrossRef]
- Balacescu, O.; Dumitrescu, R.G.; Marian, C. MicroRNAs Role in Prostate Cancer. Methods Mol. Biol. 2018, 1856, 103–117. [Google Scholar] [CrossRef]
- Shekarriz, B.; Upadhyay, J.; Wood, D.P. Intraoperative, perioperative, and long-term complications of radical prostatectomy. Urol. Clin. N. Am. 2001, 28, 639–653. [Google Scholar] [CrossRef]
- Kong, D.; Banerjee, S.; Ahmad, A.; Li, Y.; Wang, Z.; Sethi, S.; Sarkar, F.H. Epithelial to Mesenchymal Transition Is Mechanistically Linked with Stem Cell Signatures in Prostate Cancer Cells. PLoS ONE 2010, 5, e12445. [Google Scholar] [CrossRef]
- Majid, S.; Dar, A.A.; Saini, S.; Yamamura, S.; Hirata, H.; Tanaka, Y.; Deng, G.; Dahiya, R. MicroRNA-205-directed transcriptional activation of tumor suppressor genes in prostate cancer. Cancer 2010, 116, 5637–5649. [Google Scholar] [CrossRef]
- Giangreco, A.A.; Vaishnav, A.; Wagner, D.; Finelli, A.; Fleshner, N.; Van der Kwast, T.; Vieth, R.; Nonn, L. Tumor Suppressor microRNAs, miR-100 and -125b, Are Regulated by 1,25-dihydroxyvitamin D in Primary Prostate Cells and in Patient Tissue. Cancer Prev. Res. 2013, 6, 483–494. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.; Lu, Y.; Cui, D.; Li, E.; Zhu, Y.; Zhao, Y.; Zhao, F.; Xia, S. miR-200b suppresses cell proliferation, migration and enhances chemosensitivity in prostate cancer by regulating Bmi-1. Oncol. Rep. 2014, 31, 910–918. [Google Scholar] [CrossRef] [Green Version]
- Jiang, S.; Mo, C.; Guo, S.; Zhuang, J.; Huang, B.; Mao, X. Human bone marrow mesenchymal stem cells-derived microRNA-205-containing exosomes impede the progression of prostate cancer through suppression of RHPN2. J. Exp. Clin. Cancer Res. 2019, 38, 495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Lieberman, R.; Pan, J.; Zhang, Q.; Du, M.; Zhang, P.; Nevalainen, M.; Kohli, M.; Shenoy, N.K.; Meng, H.; et al. miR-375 induces docetaxel resistance in prostate cancer by targeting SEC23A and YAP1. Mol. Cancer 2016, 15, 70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, L.; Fang, Y.; Xue, J.; Chen, J. Four MicroRNAs Promote Prostate Cell Proliferation with Regulation of PTEN and Its Downstream Signals In Vitro. PLoS ONE 2013, 8, e75885. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.-Y.; Su, X.-P.; Li, Y.-N.; Guo, Y.-H. MicroRNA-425-5p promotes the development of prostate cancer via targeting forkhead box J3. Eur. Rev. Med. Pharmaco 2019, 23, 547–554. [Google Scholar] [CrossRef]
- Hussain, M.; Rathkopf, D.; Liu, G.; Armstrong, A.; Kelly, W.K.; Ferrari, A.; Hainsworth, J.; Joshi, A.; Hozak, R.R.; Yang, L.; et al. A randomised non-comparative phase II trial of cixutumumab (IMC-A12) or ramucirumab (IMC-1121B) plus mitoxantrone and prednisone in men with metastatic docetaxel-pretreated castration-resistant prostate cancer. Eur. J. Cancer 2015, 51, 1714–1724. [Google Scholar] [CrossRef] [Green Version]
- Szczyrba, J.; Nolte, E.; Wach, S.; Kremmer, E.; Stohr, R.; Hartmann, A.; Wieland, W.; Wullich, B.; Grasser, F.A. Downregulation of Sec23A Protein by miRNA-375 in Prostate Carcinoma. Mol. Cancer Res. 2011, 9, 791–800. [Google Scholar] [CrossRef] [Green Version]
- Kachakova, D.; Mitkova, A.; Popov, E.; Popov, I.; Vlahova, A.; Dikov, T.; Christova, S.; Mitev, V.; Slavov, C.; Kaneva, R. Combinations of Serum Prostate-Specific Antigen and Plasma Expression Levels of let-7c, miR-30c, miR-141, and miR-375 as Potential Better Diagnostic Biomarkers for Prostate Cancer. DNA Cell Biol. 2015, 34, 189–200. [Google Scholar] [CrossRef] [Green Version]
- Amir, S.; Ma, A.-H.; Shi, X.-B.; Xue, L.; Kung, H.-J.; deVere White, R.W. Oncomir miR-125b Suppresses p14ARF to Modulate p53-Dependent and p53-Independent Apoptosis in Prostate Cancer. PLoS ONE 2013, 8, e61064. [Google Scholar] [CrossRef]
- Shi, X.-B.; Xue, L.; Ma, A.-H.; Tepper, C.G.; Kung, H.-J.; White, R.W.D. miR-125b promotes growth of prostate cancer xenograft tumor through targeting pro-apoptotic genes. Prostate 2011, 71, 538–549. [Google Scholar] [CrossRef] [Green Version]
- Zheng, X.-M.; Zhang, P.; Liu, M.-H.; Chen, P.; Zhang, W.-B. MicroRNA-30e inhibits adhesion, migration, invasion and cell cycle progression of prostate cancer cells via inhibition of the activation of the MAPK signaling pathway by downregulating CHRM3. Int. J. Oncol. 2018. [Google Scholar] [CrossRef] [Green Version]
- Ma, Z.; Chao, F.; Wang, S.; Song, Z.; Zhuo, Z.; Zhang, J.; Xu, G.; Chen, G. CTHRC1 affects malignant tumor cell behavior and is regulated by miR-30e-5p in human prostate cancer. Biochem. Biophys. Res. Commun. 2020, 525, 418–424. [Google Scholar] [CrossRef] [PubMed]
- Bidarra, D.; Constâncio, V.; Barros-Silva, D.; Ramalho-Carvalho, J.; Moreira-Barbosa, C.; Antunes, L.; Maurício, J.; Oliveira, J.; Henrique, R.; Jerónimo, C. Circulating MicroRNAs as Biomarkers for Prostate Cancer Detection and Metastasis Development Prediction. Front. Oncol. 2019, 9, 900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, P.; Ma, L.; Zhou, J.; Jiang, M.; Rao, E.; Zhao, Y.; Guo, F. miR-17-92 plays an oncogenic role and conveys chemo-resistance to cisplatin in human prostate cancer cells. Int. J. Oncol. 2016, 48, 1737–1748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- deVere White, R.W.; Vinall, R.L.; Tepper, C.G.; Shi, X.-B. MicroRNAs and their potential for translation in prostate cancer. Urol. Oncol. Semin. Ori. 2009, 27, 307–311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiosea, S.; Jelezcova, E.; Chandran, U.; Acquafondata, M.; McHale, T.; Sobol, R.W.; Dhir, R. Up-Regulation of Dicer, a Component of the MicroRNA Machinery, in Prostate Adenocarcinoma. Am. J. Pathol. 2006, 169, 1812–1820. [Google Scholar] [CrossRef] [Green Version]
- Pang, Y.; Young, C.Y.F.; Yuan, H. MicroRNAs and prostate cancer. ABBS 2010, 42, 363–369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cochetti, G.; Poli, G.; Guelfi, G.; Boni, A.; Egidi, M.G.; Mearini, E. Different levels of serum microRNAs in prostate cancer and benign prostatic hyperplasia: Evaluation of potential diagnostic and prognostic role. OTT 2016, 9, 7545–7553. [Google Scholar] [CrossRef] [Green Version]







| PCa | HD | ||
|---|---|---|---|
| Age | (Mean ± SD) | 61.9 ± 5.7 | 54.8 ± 3.6 |
| Range | 54–71 | 50–60 | |
| Total PSA, ng/mL | 8.8 ± 0.95 | 0.9 ± 0.1 | |
| PCa stage | T1N0M0 | 27.27% | N/A |
| T2N0M0 | 72.72% | ||
| Gleason score | 5 | 9.09% | N/A |
| 6 | 27.27% | ||
| 7 | 63.63% |
| miRNA Ratios | UE | CU | P | |||
|---|---|---|---|---|---|---|
| ddCt | p | ddCt | p | ddCt | p | |
| 125b/30e | 1.0 | ** | 0.9 | oo | −1.6 | xx |
| 375/30e | 5.0 | ** | 3.9 | oo | −1.1 | xx |
| 660/375 | −4.4 | ** | −3.2 | oo | 0.7 | x |
| 660/125b | −0.4 | ** | 1.2 | xx | ||
| 660/205 | 1.5 | ** | 1.2 | x | ||
| 205/30e | −0.9 | * | −1.6 | xx | ||
| 375/31 | 5.0 | * | 4.5 | o | ||
| 375/125b | 4.0 | * | 2.9 | oo | ||
| 375/200b | 4.4 | ** | 4.3 | oo | ||
| 375/205 | 6.0 | ** | 3.8 | oo | ||
| 660/30e | 0.6 | * | 0.6 | o | ||
| 378/19b | −0.8 | * | ||||
| 425/19b | −0.8 | ** | ||||
| 200b/30e | 0.6 | ** | ||||
| 200b/125b | −0.5 | * | ||||
| 205/31 | −1.0 | ** | ||||
| 205/125b | −2.0 | ** | ||||
| 205/200b | −1.5 | ** | ||||
| 125b/31 | 1.0 | ** | ||||
| Number of differently expressed miRNA pairs | 19 | 8 | 6 | |||
| miRNARatios | UE | CU | P | ||
|---|---|---|---|---|---|
| ddCt | p | ddCt | p | ||
| 92a/19b | 5.91 | *** | |||
| 22/92a | −5.26 | ** | 1.92 | xx | |
| 22/378a | 1.78 | x | |||
| 22/425 | 1.26 | x | |||
| 378a/92a | −4.90 | *** | |||
| 425/92a | −5.71 | *** | 0.66 | x | |
| 31/30e | −7.41 | *** | |||
| 125b/30e | −4.98 | *** | |||
| 200b/30e | −7.76 | *** | |||
| 205/30e | −7.95 | *** | |||
| 375/30e | 1.53 | *** | |||
| 660/30e | −3.76 | *** | |||
| 375/31 | 7.42 | *** | |||
| 375/125b | 5.00 | *** | |||
| 375/200b | 7.78 | *** | |||
| 375/205 | 7.97 | *** | |||
| 660/205 | 4.19 | ** | |||
| 660/375 | −3.78 | *** | |||
| Number of differently expressed miRNA pairs | 16 | 0 | 4 | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Konoshenko, M.Y.; Bryzgunova, O.E.; Lekchnov, E.A.; Amelina, E.V.; Yarmoschuk, S.V.; Pak, S.V.; Laktionov, P.P. The Influence of Radical Prostatectomy on the Expression of Cell-Free MiRNA. Diagnostics 2020, 10, 600. https://doi.org/10.3390/diagnostics10080600
Konoshenko MY, Bryzgunova OE, Lekchnov EA, Amelina EV, Yarmoschuk SV, Pak SV, Laktionov PP. The Influence of Radical Prostatectomy on the Expression of Cell-Free MiRNA. Diagnostics. 2020; 10(8):600. https://doi.org/10.3390/diagnostics10080600
Chicago/Turabian StyleKonoshenko, Maria Yu., Olga E. Bryzgunova, Evgeniy A. Lekchnov, Evgeniya V. Amelina, Sergey V. Yarmoschuk, Svetlana V. Pak, and Pavel P. Laktionov. 2020. "The Influence of Radical Prostatectomy on the Expression of Cell-Free MiRNA" Diagnostics 10, no. 8: 600. https://doi.org/10.3390/diagnostics10080600