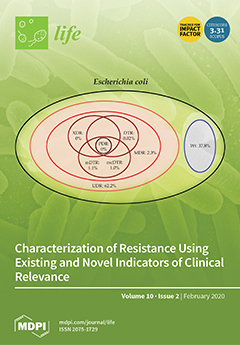
Classical resistance classifications (multidrug resistance [MDR], extensive drug resistance [XDR], pan-drug resistance [PDR]) are very useful for epidemiological purposes, however, they may not correlate well with clinical outcomes, therefore, several novel classification criteria (e.g., usual drug resistance [UDR], difficult-to-treat resistance [DTR]) were introduced
[...] Read more.
Classical resistance classifications (multidrug resistance [MDR], extensive drug resistance [XDR], pan-drug resistance [PDR]) are very useful for epidemiological purposes, however, they may not correlate well with clinical outcomes, therefore, several novel classification criteria (e.g., usual drug resistance [UDR], difficult-to-treat resistance [DTR]) were introduced for Gram-negative bacteria in recent years. Microbiological and resistance data was collected for urinary tract infections (UTIs) retrospectively, corresponding to the 2008.01.01–2017.12.31. period. Isolates were classified into various resistance categories (wild type/susceptible, UDR, MDR, XDR, DTR and PDR), in addition, two new indicators (modified DTR; mDTR and mcDTR) and a predictive composite score (pMAR) were introduced. Results: n = 16,240 (76.8%) outpatient and n = 13,386 (69.3%) inpatient UTI isolates were relevant to our analysis.
Citrobacter-Enterobacter-Serratia had the highest level of UDR isolates (88.9%), the
Proteus-Providencia-Morganella group had the highest mDTR levels. MDR levels were highest in
Acinetobacter spp. (9.7%) and
Proteus-Providencia-Morganella (9.1%). XDR- and DTR-levels were higher in non-fermenters (XDR: 1.7%–4.7%. DTR: 7.3%–7.9%) than in
Enterobacterales isolates (XDR: 0%–0.1%. DTR: 0.02%–1.5%). Conclusions: The introduction of DTR (and its’ modifications detailed in this study) to the bedside and in clinical practice will definitely lead to substantial benefits in the assessment of the significance of bacterial resistance in human therapeutics.
Full article