Differential Repeat Accumulation in the Bimodal Karyotype of Agave L.
Abstract
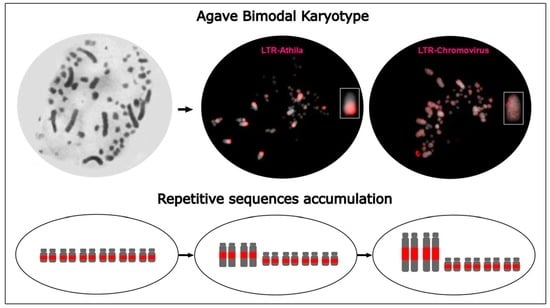
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Slide Preparation and Double Staining with CMA/DAPI
2.3. Genome Size Estimation
2.4. Sequencing of Genomic DNA and Repeat Characterization
2.5. Sequence Amplification and Fluorescent In Situ Hybridization (FISH)
2.6. Image Acquisition and Processing
3. Results
3.1. Identification and In Silico Characterization of Repetitive Sequences
3.2. Differential Distribution of Repeats along Micro- and Macrochromosomes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vosa, C.G. On chromosome uniformity, bimodality and evolution in the tribe Aloineae (Asphodelaceae). Caryologia 2005, 58, 83–85. [Google Scholar] [CrossRef] [Green Version]
- Poggio, L.; Realini, M.F.; Fourastie, M.F.; Garcia, A.M.; Gonzalez, G.E. Genome downsizing and karyotype constancy in diploid and polyploid congeners: A model of genome size variation. AoB Plants 2014, 6, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schubert, I.; Lysak, M.A. Interpretation of karyotype evolution should consider chromosome structural constraints. Trends Genet. 2011, 27, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Yin, G.S.; Yang, Z.; Chiang, T.; Gong, X. Tracing the origin of the bimodal karyotype of the tribe Lilieae (Liliaceae) based on comparative karyotypes analyses. Plant Div. Res. 2014, 36, 737–746. [Google Scholar]
- Brandham, P.E. Evolution in a stable chromosome system. In Kew Chromosome Conference II; Brandham, P.E., Bennett, M.D., Eds.; Allen and Unwin: London, UK, 1983; pp. 251–260. [Google Scholar]
- Bennett, S.T.; Kenton, A.Y.; Bennett, M.D. Genomic in situ hybridization reveals the allopolyploid nature of Milium montianum (Gramineae). Chromosoma 1992, 101, 420–424. [Google Scholar] [CrossRef]
- De la Herrán, R.; Robles, F.; Cuñado, N.; Santos, J.L.; Rejón, R.M.; Garrido-Ramos, M.A.; Rejón, R.C. A heterochromatic satellite DNA is highly amplified in a single chromosome of Muscari (Hyacinthaceae). Chromosoma 2001, 110, 197–202. [Google Scholar] [CrossRef] [PubMed]
- Báez, M.; Vaio, M.; Dreissig, S.; Schubert, V.; Houben, A.; Pedrosa-Harand, A. Together But Different: The Subgenomes of the Bimodal Eleutherine Karyotypes Are Differentially Organized. Front. Plant Sci. 2019, 10, 1170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ibiapino, A.; Báez, M.; García, M.A.; Costea, M.; Stefanović, S.; Pedrosa-Harand, A. Karyotype asymmetry in Cuscuta L. subgenus Pachystigma reflects its repeat DNA composition. Chromosome Res. 2022, 30, 91–107. [Google Scholar] [CrossRef]
- Renny-Byfield, S.; Chester, M.; Kovarik, A.; Le Comber, S.C.; Grandbastien, M.A.; Deloger, M.; Nichols, R.A.; Macas, J.; Novák, P.; Chase, M.W.; et al. Next Generation Sequencing reveals genome downsizing in allotetraploid Nicotiana tabacum, predominantly through the elimination of paternally derived repetitive DNAs. Mol. Biol. Evol. 2011, 28, 2843–2854. [Google Scholar] [CrossRef] [Green Version]
- Li, S.-F.; Su, T.; Cheng, G.-Q.; Wang, B.-X.; Li, X.; Deng, C.-L.; Gao, W.-J. Chromosome Evolution in Connection with Repetitive Sequences and Epigenetics in Plants. Genes 2017, 8, 290. [Google Scholar] [CrossRef] [Green Version]
- Kovarik, A.; Dadejova, M.; Lim, Y.K.; Chase, M.W.; Clarkson, J.J.; Knapp, S.; Leitch, A.R. Evolution of rDNA in Nicotiana allopolyploids: A potential link between rDNA homogenization and epigenetics. Ann. Bot. 2008, 101, 815–823. [Google Scholar] [CrossRef] [PubMed]
- Klein, S.J.; O’Neill, R.J. Transposable elements: Genome innovation, chromosome diversity, and centromere conflict. Chromosome Res. 2018, 26, 5–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ribeiro, T.; Vaio, M.; Félix, L.P.; Guerra, M. Satellite DNA probes of Alstroemeria longistaminea (Alstroemeriaceae) paint the heterochromatin and the B chromosome, reveal a G-like banding pattern, and point to a strong structural karyotype conservation. Protoplasma 2022, 259, 413–426. [Google Scholar] [CrossRef] [PubMed]
- Hofstatter, P.G.; Thangavel, G.; Lux, T.; Neumann, P.; Vondrak, T.; Novak, P.; Zhang, M.; Costa, C.; Castellani, M.; Scott, A.; et al. Repeat-based holocentromeres influence genome architecture and karyotype evolution. Cell 2022, 185, 3153–3168. [Google Scholar] [CrossRef] [PubMed]
- Tomaszewska, P.; Schwarzacher, T.; Heslop-Harrison, J.P. Oat chromosome and genome evolution defined by widespread terminal intergenomic translocations in polyploids. Front. Plant Sci. 2022, 13, 1026364. [Google Scholar] [CrossRef] [PubMed]
- Palomino, G.; Martínez, J.; Romero, P.; Barba-González, R.; Rodríguez-Garay, B. Nuclear genome size and karyotype analysis of Agave angustifolia Haw. “Cimarron” and “Lineño” (Asparagales, Asparagaceae). Caryologia 2017, 70, 93–101. [Google Scholar] [CrossRef]
- Granick, E.B. A karyosystematic study of the genus Agave. Am. J. Bot. 1944, 31, 283–298. [Google Scholar] [CrossRef]
- McKain, M.R.; Wickett, N.; Zhang, Y.; Ayyampalayam, S.; McCombie, W.R.; Chase, M.W.; Pires, J.C.; de Pamphilis, C.W.; Leebens-Mack, J. Phylogenomic analysis of transcriptome data elucidates co-occurrence of a paleopolyploid event and the origin of bimodal karyotypes in Agavoideae (Asparagaceae). Am. J. Bot. 2012, 99, 397–406. [Google Scholar] [CrossRef] [Green Version]
- Tamayo-Ordóñez, Y.J.; Narváez-Zapata, J.A.; Tamayo-Ordóñez, M.C.; Sánchez-Teyer, L.F. Retroelements and DNA methylation could contribute to diversity of 5S rDNA in Agave L. J. Mol. Evol. 2018, 86, 404–423. [Google Scholar] [CrossRef]
- Good-Avila, S.V.; Souza, V.; Gaut, B.S.; Eguiarte, L.E. Timing and rate of speciation in Agave (Agavaceae). Proc. Natl. Acad. Sci. USA 2006, 103, 9124–9129. [Google Scholar] [CrossRef] [Green Version]
- Hulle, A.; Kadole, P.; Katkar, P. Agave Americana leaf fibers. Fibers 2015, 3, 64–75. [Google Scholar] [CrossRef]
- Monja-Mio, K.M.; Herrera-Alamillo, M.A.; Sánchez-Teyer, L.F.; Robert, M.L. Breeding strategies to improve production of Agave (Agave spp.). In Advances in Plant Breeding Strategies: Industrial and Food Crops; Al-Khayri, J., Jain, S., Johnson, D., Eds.; Springer International Publishing: Cham, Switzerland, 2019; Volume 6, pp. 319–362. [Google Scholar] [CrossRef]
- Huang, X.; Xiao, M.; Xi, J.; He, C.; Zheng, J.; Chen, H.; Gao, J.; Zhang, S.; Wu, W.; Liang, Y.; et al. De Novo transcriptome assembly of Agave H11648 by Illumina sequencing and identification of cellulose synthase genes in Agave species. Genes 2019, 102, 103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Queiroz, S.R.O.D.; Ortolani, F.A.; Mataqueiro, M.F.; Osuna, J.T.A.; Moro, J.R. Análise cromossômica em bulbilhos de sisal (Agave spp.) cultivados em diferentes municípios baianos, Brasil. Acta Bot. Bras. 2012, 26, 842–848. [Google Scholar] [CrossRef] [Green Version]
- Robert, M.L.; Lim, K.Y.; Hanson, L.; Sanchez-Teyer, F.; Bennett, M.D.; Leitch, A.R.; Leitch, I.J. Wild and agronomically important Agave species (Asparagaceae) show proportional increases in chromosome number, genome size, and genetic markers with increasing ploidy. Bot. J. Linn. Soc. 2008, 158, 215–222. [Google Scholar] [CrossRef] [Green Version]
- Doughty, L.R. Chromosome behaviour in relation to genetics of Agave. J. Genet. 1936, 33, 198–205. [Google Scholar] [CrossRef]
- Trejo-Torres, J.C.; Gann, G.D.; Christenhusz, M.J. The Yucatan Peninsula is the place of origin of sisal (Agave sisalana, Asparagaceae): Historical accounts, phytogeography and current populations. Bot. Sci. 2018, 96, 366–379. [Google Scholar] [CrossRef] [Green Version]
- Finch, R.A.; Osborne, J.F. Chromosome numbers and DNA amounts in Agave variants. East Afr. Agric. For. J. 1990, 55, 213–218. [Google Scholar] [CrossRef]
- Moreno-Salazar, S.F.; Esqueda, M.; Martínez, J.; Palomino, G. Tamaño del genoma y cariotipo en Agave angustifolia y A. rhodacantha de Sonora, México. Rev. Fitotec. Mex. 2007, 30, 13–23. [Google Scholar] [CrossRef]
- Gomez-Rodriguez, V.M.; Rodriguez-Garay, B.; Palomino, G.; Martínez, J.; Barba-Gonzalez, R. Physical mapping of 5S and 18S ribosomal DNA in three species of Agave (Asparagales, Asparagaceae). Comp. Cytogen 2013, 7, 191–203. [Google Scholar] [CrossRef]
- Jiménez-Barron, O.; García-Sandoval, R.; Magallón, S.; García-Mendoza, A.; Nieto-Sotelo, J.; Aguirre-Planter, E.; Eguiarte, L.E. Phylogeny, diversification rate, and divergence time of Agave sensu lato (Asparagaceae), a group of recent origin in the process of diversification. Front. Plant Sci. 2020, 11, 536135. [Google Scholar] [CrossRef]
- Xu, B.; Tan, S.; Qin, X.; Huang, X.; Xi, J.; Chen, H.; Qin, J.; Chen, T.; Yi, K. The complete chloroplast genome of Agave amaniensis (Asparagales: Asparagaceae: Agavoideae). Mitochondrial DNA B Resour. 2022, 7, 1519–1521. [Google Scholar] [CrossRef] [PubMed]
- De Carvalho, C.R.; Saraiva, L.S. An Air Drying Technique for Maize Chromosomes without Enzymatic Maceration. Biotech. Histochem. 1993, 68, 142–145. [Google Scholar] [CrossRef] [PubMed]
- Schweizer, D.; Ambros, P.E. Chromosome Banding. In Chromosome Analysis Protocols. Methods in Molecular Biology™; Gosden, J.R., Ed.; Humana Press. Inc.: Totowa, NJ, USA, 1994; Volume 29, pp. 97–112. ISBN 978-1-59259-516-7. [Google Scholar] [CrossRef]
- Doležel, J.; Greilhuber, J.; Suda, J. Estimation of nuclear DNA content in plants using flow cytometry. Nat. Protoc. 2007, 2, 2233. [Google Scholar] [CrossRef] [PubMed]
- Novák, P.; Neumann, P.; Macas, J. Graph-based clustering and characterization of repetitive sequences in next-generation sequencing data. BMC Bioinform. 2010, 11, 378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Novak, P.; Neumann, P.; Pech, J.; Steinhaisl, J.; Macas, J. RepeatExplorer: A Galaxy-based web server for genome-wide characterization of eukaryotic repetitive elements from next-generation sequence reads. Bioinformatics 2013, 29, 792–793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sonnhammer, E.L.L.; Durbin, R. A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene 1995, 167, GC1–GC10. [Google Scholar] [CrossRef] [Green Version]
- Neumann, P.; Novák, P.; Hoštáková, N.; Macas, J. Systematic survey of plant LTR-retrotransposons elucidates phylogenetic relationships of their polyprotein domains and provides a reference for element classification. Mob DNA 2019, 10, 1. [Google Scholar] [CrossRef]
- Pedrosa, A.; Sandal, N.; Stougaard, J.; Schweizer, D.; Bachmair, A. Chromosomal map of the model legume Lotus japonicus. Genetics 2002, 161, 1661–1672. [Google Scholar] [CrossRef] [PubMed]
- Wanzenbock, E.-M.; Schofer, C.; Schweizer, D.; Bachmair, A. Ribosomal transcription units integrated via T-DNA transformation associate with the nucleolus and do not require upstream repeat sequences for activity in Arabidopsis thaliana. Plant J. 1997, 11, 1007–1016. [Google Scholar] [CrossRef]
- Hertweck, K.L. Assembly and comparative analysis of transposable elements from low coverage genomic sequence data in Asparagales. Genome 2013, 56, 487–494. [Google Scholar] [CrossRef] [PubMed]
- Khaliq, I.; Khan, M.A.; Pearce, S. Ty1-Copia retrotransposons are heterogeneous, extremely high copy number and are major players in the genome organization and evolution of Agave tequilana. Genet. Resour. Crop. Evol. 2011, 59, 575–587. [Google Scholar] [CrossRef]
- Espinosa-Barrera, L.A.; Sánchez-Teyer, L.F.; Quiroz-Moreno, A.; Narváez-Zapata, J.A. Identification and characterization of a new satellite-like DNA family in three Agave species. Plant Gene 2018, 16, 8–18. [Google Scholar] [CrossRef]
- Sader, M.; Vaio, M.; Cauz-Santos, L.A.; Dornelas, M.C.; Vieira, M.L.C.; Melo, N.; Pedrosa-Harand, A. Large vs small genomes in Passiflora: The influence of the mobilome and the satellitome. Planta 2021, 253, 86. [Google Scholar] [CrossRef] [PubMed]
- Van-Lume, B.; Mata-Sucre, Y.; Báez, M.; Ribeiro, T.; Huettel, B.; Gagnon, E.; Leitch, I.J.; Pedrosa-Harand, A.; Lewis, G.P.; Souza, G. Evolutionary convergence or homology? Comparative cytogenomics of Caesalpinia group species (Leguminosae) reveals diversification in the pericentromeric heterochromatic composition. Planta 2019, 250, 2173–2186. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, T.; Vasconcelos, E.; Dos Santos, K.G.; Vaio, M.; Brasileiro-Vidal, A.C.; Pedrosa-Harand, A. Diversity of repetitive sequences within compact genomes of Phaseolus L. beans and allied genera Cajanus L. and Vigna Savi. Chromosome Res. 2020, 28, 139–153. [Google Scholar] [CrossRef] [PubMed]
- Melters, D.P.; Bradnam, K.R.; Young, H.A.; Telis, N.; May, M.R.; Ruby, J.G.; Sebra, R.; Peluso, P.; Eid, J.; Rank, D.; et al. Comparative analysis of tandem repeats from hundreds of species reveals unique insights into centromere evolution. Genome Biol. 2013, 14, R10. [Google Scholar] [CrossRef]
- Ruiz-Ruano, F.J.; López-León, M.D.; Cabrero, J.; Camacho, J.P.M. High-throughput analysis of the satellitome illuminates satellite DNA evolution. Sci. Rep. 2016, 6, 28333. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Zuo, S.; Li, Z.; Meng, Z.; Han, J.; Song, J.; Pan, Y.-B.; Wang, K. Isolation and characterization of centromeric repetitive DNA sequences in Saccharum spontaneum. Sci. Rep. 2017, 7, 41659. [Google Scholar] [CrossRef] [Green Version]
- Whitaker, T.W. Chromosome constitution in certain monocotyledons. J. Arnold Arbor. 1934, 15, 135–153. [Google Scholar] [CrossRef]


| Repetitive Elements | Genome % | |
|---|---|---|
| Retrotransposons | 64.32 | |
| SINE | 0.46 | |
| LTRs | 62.50 | |
| LTR non classified | 20.99 | |
| Ty1/copia | 24.90 | |
| SIRE | 4.00 | |
| Tork | 1.92 | |
| TAR | 1.41 | |
| Angela | 0.90 | |
| Bianca | 0.17 | |
| Ivana/Oryco | 0.16 | |
| Ale I | 0.12 | |
| Ale II | 0.12 | |
| Unclassified Ty1/copia | 16.12 | |
| Ty3/gypsy | 16.61 | |
| Ogre | 3.36 | |
| Athila | 2.37 | |
| Chromovirus | 1.97 | |
| Unclassified Ty3/gypsy | 8.92 | |
| rDNA | 0.12 | |
| DNA Transposon | 0.58 | |
| Satellite DNA | 1.99 | |
| Unclassified | 0.68 | |
| Total genome proportion | 67.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramos, L.C.; Báez, M.; Fuchs, J.; Houben, A.; Carvalho, R.; Pedrosa-Harand, A. Differential Repeat Accumulation in the Bimodal Karyotype of Agave L. Genes 2023, 14, 491. https://doi.org/10.3390/genes14020491
Ramos LC, Báez M, Fuchs J, Houben A, Carvalho R, Pedrosa-Harand A. Differential Repeat Accumulation in the Bimodal Karyotype of Agave L. Genes. 2023; 14(2):491. https://doi.org/10.3390/genes14020491
Chicago/Turabian StyleRamos, Lamonier Chaves, Mariana Báez, Joerg Fuchs, Andreas Houben, Reginaldo Carvalho, and Andrea Pedrosa-Harand. 2023. "Differential Repeat Accumulation in the Bimodal Karyotype of Agave L." Genes 14, no. 2: 491. https://doi.org/10.3390/genes14020491