The Potential of ANK1 to Predict Parkinson’s Disease
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microarray Data Information
2.2. Animals and Reagents
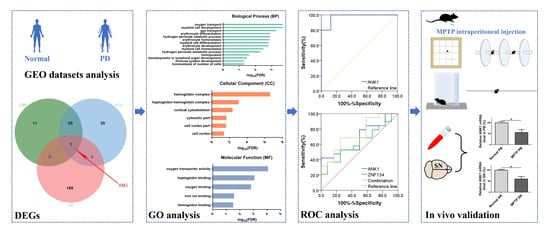
2.3. DEGs Assessment
2.4. Protein–Protein Interaction Analysis
2.5. Genome Ontology (GO) Enrichment Analysis
2.6. Receiver Operating Characteristics (ROC) Curve Analysis
2.7. Construction of MPTP-Induced PD Model
2.8. Behavioral Tests
2.9. Polymerase Chain Reaction (PCR) Analysis
2.10. Western Blot Analysis
2.11. Immunofluorescence
2.12. In Vitro Cell Viability Assay
2.13. Statistical Analysis
3. Results
3.1. Identification of ANK1 as a Commonly Down-Regulated Gene
3.2. Construction of PD Associated Regulatory Network and Module Analysis
3.3. ANK1 Confers Significant Correlation with PD Progression
3.4. ANK1 Shows a High Diagnosis Capacity in PD
3.5. Validation of ANK1 in MPTP-Induced PD Model
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gui, C.; Ren, Y.; Chen, J.; Wu, X.; Mao, K.; Li, H.; Yu, H.; Zou, F.; Li, W. p38 MAPK-DRP1 signaling is involved in mitochondrial dysfunction and cell death in mutant A53T α-synuclein model of Parkinson’s disease. Toxicol. Appl. Pharmacol. 2020, 388, 114874. [Google Scholar] [CrossRef]
- Ikeda-Matsuo, Y.; Miyata, H.; Mizoguchi, T.; Ohama, E.; Naito, Y.; Uematsu, S.; Akira, S.; Sasaki, Y.; Tanabe, M. Microsomal prostaglandin E synthase-1 is a critical factor in dopaminergic neurodegeneration in Parkinson’s disease. Neurobiol. Dis. 2019, 124, 81–92. [Google Scholar] [CrossRef]
- Zaafan, M.A.; Abdelhamid, A.M.; Ibrahim, S.M. The Protective Effect of Korean Red Ginseng Against Rotenone-Induced Parkinson’s Disease in Rat Model: Modulation of Nuclear Factor-κβ and Caspase-3. Curr. Pharm. Biotechnol. 2019, 20, 588–594. [Google Scholar] [CrossRef]
- Guo, J.D.; Zhao, X.; Li, Y.; Li, G.R.; Liu, X.L. Damage to dopaminergic neurons by oxidative stress in Parkinson’s disease (Review). Int. J. Mol. Med. 2018, 41, 1817–1825. [Google Scholar] [CrossRef] [Green Version]
- Shukla, L.; Yuan, Y.; Shayan, R.; Greening, D.W.; Karnezis, T. Fat Therapeutics: The Clinical Capacity of Adipose-Derived Stem Cells and Exosomes for Human Disease and Tissue Regeneration. Front. Pharmacol. 2020, 11, 158. [Google Scholar] [CrossRef] [Green Version]
- Haikal, C.; Chen, Q.Q.; Li, J.Y. Microbiome changes: An indicator of Parkinson’s disease? Transl. Neurodegener. 2019, 8, 38. [Google Scholar] [CrossRef] [Green Version]
- Charvin, D.; Medori, R.; Hauser, R.A.; Rascol, O. Therapeutic strategies for Parkinson disease: Beyond dopaminergic drugs. Nat. Rev. Drug Discov. 2018, 17, 804–822. [Google Scholar] [CrossRef]
- Kasai, S.; Yoshihara, T.; Lopatina, O.; Ishihara, K.; Higashida, H. Selegiline Ameliorates Depression-Like Behavior in Mice Lacking the CD157/BST1 Gene, a Risk Factor for Parkinson’s Disease. Front. Behav. Neurosci. 2017, 11, 75. [Google Scholar] [CrossRef]
- Pasluosta, C.F.; Steib, S.; Klamroth, S.; Gaßner, H.; Goßler, J.; Hannink, J.; von Tscharner, V.; Pfeifer, K.; Winkler, J.; Klucken, J.; et al. Acute Neuromuscular Adaptations in the Postural Control of Patients with Parkinson’s Disease after Perturbed Walking. Front. Aging Neurosci. 2017, 9, 316. [Google Scholar] [CrossRef]
- Yang, P.; Perlmutter, J.S.; Benzinger, T.L.S.; Morris, J.C.; Xu, J. Dopamine D3 receptor: A neglected participant in Parkinson Disease pathogenesis and treatment? Ageing Res. Rev. 2020, 57, 100994. [Google Scholar] [CrossRef]
- Kalia, L.V.; Lang, A.E. Parkinson’s disease. Lancet (Lond. Engl.) 2015, 386, 896–912. [Google Scholar] [CrossRef]
- Milanowski, Ł.M.; Ross, O.A.; Friedman, A.; Hoffman-Zacharska, D.; Gorka-Skoczylas, P.; Jurek, M.; Koziorowski, D.; Wszolek, Z.K. Genetics of Parkinson’s disease in the Polish population. Neurol. I Neurochir. Pol. 2021, 55, 241–252. [Google Scholar] [CrossRef]
- Nalls, M.A.; Blauwendraat, C.; Vallerga, C.L.; Heilbron, K.; Bandres-Ciga, S.; Chang, D.; Tan, M.; Kia, D.A.; Noyce, A.J.; Xue, A.; et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: A meta-analysis of genome-wide association studies. Lancet. Neurol. 2019, 18, 1091–1102. [Google Scholar] [CrossRef]
- Sancandi, M.; Schul, E.V.; Economides, G.; Constanti, A.; Mercer, A. Structural Changes Observed in the Piriform Cortex in a Rat Model of Pre-motor Parkinson’s Disease. Front. Cell. Neurosci. 2018, 12, 479. [Google Scholar] [CrossRef] [Green Version]
- Armstrong, M.J.; Okun, M.S. Diagnosis and Treatment of Parkinson Disease: A Review. JAMA 2020, 323, 548–560. [Google Scholar] [CrossRef]
- Nizamani, W.M.; Mubarak, F.; Barakzai, M.D.; Ahmed, M.S. Role of magnetic resonance planimetry and magnetic resonance parkinsonism index in discriminating Parkinson’s disease and progressive supranuclear palsy: A retrospective study based on 1.5 and 3 T MRI. Int. J. Gen. Med. 2017, 10, 375–384. [Google Scholar] [CrossRef] [Green Version]
- Postuma, R.B.; Berg, D.; Stern, M.; Poewe, W.; Olanow, C.W.; Oertel, W.; Obeso, J.; Marek, K.; Litvan, I.; Lang, A.E.; et al. MDS clinical diagnostic criteria for Parkinson’s disease. Mov. Disord. Off. J. Mov. Disord. Soc. 2015, 30, 1591–1601. [Google Scholar] [CrossRef]
- Suwijn, S.R.; van Boheemen, C.J.; de Haan, R.J.; Tissingh, G.; Booij, J.; de Bie, R.M. The diagnostic accuracy of dopamine transporter SPECT imaging to detect nigrostriatal cell loss in patients with Parkinson’s disease or clinically uncertain parkinsonism: A systematic review. EJNMMI Res. 2015, 5, 12. [Google Scholar] [CrossRef] [Green Version]
- Tolosa, E.; Wenning, G.; Poewe, W. The diagnosis of Parkinson’s disease. Lancet Neurol. 2006, 5, 75–86. [Google Scholar] [CrossRef]
- Xing, Y.; Sapuan, A.; Dineen, R.A.; Auer, D.P. Life span pigmentation changes of the substantia nigra detected by neuromelanin-sensitive MRI. Mov. Disord. Off. J. Mov. Disord. Soc. 2018, 33, 1792–1799. [Google Scholar] [CrossRef]
- Poewe, W.; Seppi, K.; Tanner, C.M.; Halliday, G.M.; Brundin, P.; Volkmann, J.; Schrag, A.E.; Lang, A.E. Parkinson disease. Nat. Rev. Dis. Prim. 2017, 3, 17013. [Google Scholar] [CrossRef]
- Locascio, J.J.; Eberly, S.; Liao, Z.; Liu, G.; Hoesing, A.N.; Duong, K.; Trisini-Lipsanopoulos, A.; Dhima, K.; Hung, A.Y.; Flaherty, A.W.; et al. Association between α-synuclein blood transcripts and early, neuroimaging-supported Parkinson’s disease. Brain A J. Neurol. 2015, 138 Pt 9, 2659–2671. [Google Scholar] [CrossRef]
- Starhof, C.; Hejl, A.M.; Heegaard, N.H.H.; Carlsen, A.L.; Burton, M.; Lilje, B.; Winge, K. The biomarker potential of cell-free microRNA from cerebrospinal fluid in Parkinsonian Syndromes. Mov. Disord. Off. J. Mov. Disord. Soc. 2019, 34, 246–254. [Google Scholar] [CrossRef]
- Santiago, J.A.; Potashkin, J.A. Blood Transcriptomic Meta-analysis Identifies Dysregulation of Hemoglobin and Iron Metabolism in Parkinson’ Disease. Front. Aging Neurosci. 2017, 9, 73. [Google Scholar] [CrossRef] [Green Version]
- Shi, M.; Zabetian, C.P.; Hancock, A.M.; Ginghina, C.; Hong, Z.; Yearout, D.; Chung, K.A.; Quinn, J.F.; Peskind, E.R.; Galasko, D.; et al. Significance and confounders of peripheral DJ-1 and alpha-synuclein in Parkinson’s disease. Neurosci. Lett. 2010, 480, 78–82. [Google Scholar] [CrossRef] [Green Version]
- Yazdani, U.; Zaman, S.; Hynan, L.S.; Brown, L.S.; Dewey, R.B., Jr.; Karp, D.; German, D.C. Blood biomarker for Parkinson disease: Peptoids. NPJ Park. Dis. 2016, 2, 16012. [Google Scholar] [CrossRef] [Green Version]
- Cilento, E.M.; Jin, L.; Stewart, T.; Shi, M.; Sheng, L.; Zhang, J. Mass spectrometry: A platform for biomarker discovery and validation for Alzheimer’s and Parkinson’s diseases. J. Neurochem. 2019, 151, 397–416. [Google Scholar] [CrossRef]
- Tessema, M.; Yingling, C.M.; Picchi, M.A.; Wu, G.; Ryba, T.; Lin, Y.; Bungum, A.O.; Edell, E.S.; Spira, A.; Belinsky, S.A. ANK1 Methylation regulates expression of MicroRNA-486-5p and discriminates lung tumors by histology and smoking status. Cancer Lett. 2017, 410, 191–200. [Google Scholar] [CrossRef]
- Zhou, D.; Birkenmeier, C.S.; Williams, M.W.; Sharp, J.J.; Barker, J.E.; Bloch, R.J. Small, membrane-bound, alternatively spliced forms of ankyrin 1 associated with the sarcoplasmic reticulum of mammalian skeletal muscle. J. Cell Biol. 1997, 136, 621–631. [Google Scholar] [CrossRef]
- Makara, M.A.; Curran, J.; Little, S.C.; Musa, H.; Polina, I.; Smith, S.A.; Wright, P.J.; Unudurthi, S.D.; Snyder, J.; Bennett, V.; et al. Ankyrin-G coordinates intercalated disc signaling platform to regulate cardiac excitability in vivo. Circ. Res. 2014, 115, 929–938. [Google Scholar] [CrossRef]
- Chai, S.; Jiao, R.; Sun, X.; Fu, P.; Zhao, Q.; Sang, M. Novel nonsense mutation p. Gln264Ter in the ANK1 confirms causative role for hereditary spherocytosis: A case report. BMC Med. Genet. 2020, 21, 223. [Google Scholar] [CrossRef] [PubMed]
- Imamura, M.; Maeda, S.; Yamauchi, T.; Hara, K.; Yasuda, K.; Morizono, T.; Takahashi, A.; Horikoshi, M.; Nakamura, M.; Fujita, H.; et al. A single-nucleotide polymorphism in ANK1 is associated with susceptibility to type 2 diabetes in Japanese populations. Hum. Mol. Genet. 2012, 21, 3042–3049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harder, M.N.; Ribel-Madsen, R.; Justesen, J.M.; Sparsø, T.; Andersson, E.A.; Grarup, N.; Jørgensen, T.; Linneberg, A.; Hansen, T.; Pedersen, O. Type 2 diabetes risk alleles near BCAR1 and in ANK1 associate with decreased β-cell function whereas risk alleles near ANKRD55 and GRB14 associate with decreased insulin sensitivity in the Danish Inter99 cohort. J. Clin. Endocrinol. Metab. 2013, 98, E801–E806. [Google Scholar] [CrossRef] [Green Version]
- Lunnon, K.; Smith, R.; Hannon, E.; De Jager, P.L.; Srivastava, G.; Volta, M.; Troakes, C.; Al-Sarraj, S.; Burrage, J.; Macdonald, R.; et al. Methylomic profiling implicates cortical deregulation of ANK1 in Alzheimer’s disease. Nat. Neurosci. 2014, 17, 1164–1170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desplats, P.; Gutierrez, A.M.; Antonelli, M.C.; Frasch, M.G. Microglial memory of early life stress and inflammation: Susceptibility to neurodegeneration in adulthood. Neurosci. Biobehav. Rev. 2020, 117, 232–242. [Google Scholar] [CrossRef] [Green Version]
- Smith, A.R.; Smith, R.G.; Burrage, J.; Troakes, C.; Al-Sarraj, S.; Kalaria, R.N.; Sloan, C.; Robinson, A.C.; Mill, J.; Lunnon, K. A cross-brain regions study of ANK1 DNA methylation in different neurodegenerative diseases. Neurobiol. Aging 2019, 74, 70–76. [Google Scholar] [CrossRef] [Green Version]
- Plowman, E.K.; Maling, N.; Rivera, B.J.; Larson, K.; Thomas, N.J.; Fowler, S.C.; Manfredsson, F.P.; Shrivastav, R.; Kleim, J.A. Differential sensitivity of cranial and limb motor function to nigrostriatal dopamine depletion. Behav. Brain Res. 2013, 237, 157–163. [Google Scholar] [CrossRef] [Green Version]
- Zhu, M.; Gong, D. A Mouse Model of 1-Methyl-4-Phenyl-1,2,3,6-Tetrahydropyridine (MPTP)-Induced Parkinson Disease Shows that 2-Aminoquinoline Targets JNK Phosphorylation. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2020, 26, e920989. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, A.; Huang, J. Identification of Gene Changes Induced by Dexamethasone in the Anterior Segment of the Human Eye Using Bioinformatics Analysis. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2019, 25, 5501–5509. [Google Scholar] [CrossRef]
- Bernhard, F.P.; Heinzel, S.; Binder, G.; Weber, K.; Apel, A.; Roeben, B.; Deuschle, C.; Maechtel, M.; Heger, T.; Nussbaum, S.; et al. Insulin-Like Growth Factor 1 (IGF-1) in Parkinson’s Disease: Potential as Trait-, Progression- and Prediction Marker and Confounding Factors. PLoS ONE 2016, 11, e0150552. [Google Scholar] [CrossRef]
- Xiong, Y.; You, W.; Wang, R.; Peng, L.; Fu, Z. Prediction and Validation of Hub Genes Associated with Colorectal Cancer by Integrating PPI Network and Gene Expression Data. BioMed Res. Int. 2017, 2017, 2421459. [Google Scholar] [CrossRef] [Green Version]
- Pihlstrøm, L.; Blauwendraat, C.; Cappelletti, C.; Berge-Seidl, V.; Langmyhr, M.; Henriksen, S.P.; van de Berg, W.D.J.; Gibbs, J.R.; Cookson, M.R.; Singleton, A.B.; et al. A comprehensive analysis of SNCA-related genetic risk in sporadic parkinson disease. Ann. Neurol. 2018, 84, 117–129. [Google Scholar] [CrossRef] [PubMed]
- Potashkin, J.A.; Santiago, J.A.; Ravina, B.M.; Watts, A.; Leontovich, A.A. Biosignatures for Parkinson’s disease and atypical parkinsonian disorders patients. PLoS ONE 2012, 7, e43595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guizouarn, H.; Martial, S.; Gabillat, N.; Borgese, F. Point mutations involved in red cell stomatocytosis convert the electroneutral anion exchanger 1 to a nonselective cation conductance. Blood 2007, 110, 2158–2165. [Google Scholar] [CrossRef] [PubMed]
- Shehadeh, L.A.; Yu, K.; Wang, L.; Guevara, A.; Singer, C.; Vance, J.; Papapetropoulos, S. SRRM2, a potential blood biomarker revealing high alternative splicing in Parkinson’s disease. PLoS ONE 2010, 5, e9104. [Google Scholar] [CrossRef]
- Medlock, A.; Swartz, L.; Dailey, T.A.; Dailey, H.A.; Lanzilotta, W.N. Substrate interactions with human ferrochelatase. Proc. Natl. Acad. Sci. USA 2007, 104, 1789–1793. [Google Scholar] [CrossRef] [Green Version]
- Santiago, J.A.; Potashkin, J.A. Network-based metaanalysis identifies HNF4A and PTBP1 as longitudinally dynamic biomarkers for Parkinson’s disease. Proc. Natl. Acad. Sci. USA 2015, 112, 2257–2262. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Yu, W.; Li, Y.; Shang, X.; Zhang, X.; Xiong, F.; Xu, X. Analysis of alpha-hemoglobin-stabilizing protein (AHSP) gene as a genetic modifier to the phenotype of beta-thalassemia in Southern China. Blood Cells Mol. Dis. 2010, 45, 128–132. [Google Scholar] [CrossRef]
- Burgess-Herbert, S.L.; Tsaih, S.W.; Stylianou, I.M.; Walsh, K.; Cox, A.J.; Paigen, B. An experimental assessment of in silico haplotype association mapping in laboratory mice. BMC Genet. 2009, 10, 81. [Google Scholar] [CrossRef] [Green Version]
- Shankar, J.; Cerqueira, G.C.; Wortman, J.R.; Clemons, K.V.; Stevens, D.A. RNA-Seq Profile Reveals Th-1 and Th-17-Type of Immune Responses in Mice Infected Systemically with Aspergillus fumigatus. Mycopathologia 2018, 183, 645–658. [Google Scholar] [CrossRef]
- Elhodaky, M.; Diamond, A.M. Selenium-Binding Protein 1 in Human Health and Disease. Int. J. Mol. Sci. 2018, 19, 3437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor, M.W.; Tsukahara, T.; McClintick, J.N.; Edenberg, H.J.; Kwo, P. Cyclic changes in gene expression induced by Peg-interferon alfa-2b plus ribavirin in peripheral blood monocytes (PBMC) of hepatitis C patients during the first 10 weeks of treatment. J. Transl. Med. 2008, 6, 66. [Google Scholar] [CrossRef] [Green Version]
- Schechter, A.N. Hemoglobin research and the origins of molecular medicine. Blood 2008, 112, 3927–3938. [Google Scholar] [CrossRef] [Green Version]
- Abbott, R.D.; Ross, G.W.; Tanner, C.M.; Andersen, J.K.; Masaki, K.H.; Rodriguez, B.L.; White, L.R.; Petrovitch, H. Late-life hemoglobin and the incidence of Parkinson’s disease. Neurobiol. Aging 2012, 33, 914–920. [Google Scholar] [CrossRef] [Green Version]
- Savica, R.; Grossardt, B.R.; Carlin, J.M.; Icen, M.; Bower, J.H.; Ahlskog, J.E.; Maraganore, D.M.; Steensma, D.P.; Rocca, W.A. Anemia or low hemoglobin levels preceding Parkinson disease: A case-control study. Neurology 2009, 73, 1381–1387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ward, R.J.; Zucca, F.A.; Duyn, J.H.; Crichton, R.R.; Zecca, L. The role of iron in brain ageing and neurodegenerative disorders. Lancet. Neurol. 2014, 13, 1045–1060. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Huang, X.; Wang, J.; Huang, R.; Wan, D. Regulation of Iron Homeostasis and Related Diseases. Mediat. Inflamm. 2020, 2020, 6062094. [Google Scholar] [CrossRef]
- Sengstock, G.J.; Olanow, C.W.; Menzies, R.A.; Dunn, A.J.; Arendash, G.W. Infusion of iron into the rat substantia nigra: Nigral pathology and dose-dependent loss of striatal dopaminergic markers. J. Neurosci. Res. 1993, 35, 67–82. [Google Scholar] [CrossRef]
- Gerlach, M.; Ben-Shachar, D.; Riederer, P.; Youdim, M.B. Altered brain metabolism of iron as a cause of neurodegenerative diseases? J. Neurochem. 1994, 63, 793–807. [Google Scholar] [CrossRef]
- Pichler, I.; Del Greco, M.F.; Gögele, M.; Lill, C.M.; Bertram, L.; Do, C.B.; Eriksson, N.; Foroud, T.; Myers, R.H.; Nalls, M.; et al. Serum iron levels and the risk of Parkinson disease: A Mendelian randomization study. PLoS Med. 2013, 10, e1001462. [Google Scholar] [CrossRef]
- Miyake, Y.; Tanaka, K.; Fukushima, W.; Sasaki, S.; Kiyohara, C.; Tsuboi, Y.; Yamada, T.; Oeda, T.; Miki, T.; Kawamura, N.; et al. Dietary intake of metals and risk of Parkinson’s disease: A case-control study in Japan. J. Neurol. Sci. 2011, 306, 98–102. [Google Scholar] [CrossRef]
- Chen, J.J.; Zhang, S. Heme-regulated eIF2α kinase in erythropoiesis and hemoglobinopathies. Blood 2019, 134, 1697–1707. [Google Scholar] [CrossRef] [PubMed]
- Camaschella, C. Iron deficiency. Blood 2019, 133, 30–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, F.; Wu, Q.; Sun, S.; Bi, G.; Guo, L. Identification of potential diagnostic biomarkers for Parkinson’s disease. FEBS Open Bio 2019, 9, 1460–1468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soreq, L.; Israel, Z.; Bergman, H.; Soreq, H. Advanced microarray analysis highlights modified neuro-immune signaling in nucleated blood cells from Parkinson’s disease patients. J. Neuroimmunol. 2008, 201–202, 227–236. [Google Scholar] [CrossRef]
- Sakharkar, M.K.; Kashmir Singh, S.K.; Rajamanickam, K.; Mohamed Essa, M.; Yang, J.; Chidambaram, S.B. A systems biology approach towards the identification of candidate therapeutic genes and potential biomarkers for Parkinson’s disease. PLoS ONE 2019, 14, e0220995. [Google Scholar] [CrossRef] [Green Version]
- Santiago, J.A.; Scherzer, C.R.; Potashkin, J.A. Specific splice variants are associated with Parkinson’s disease. Mov. Disord. Off. J. Mov. Disord. Soc. 2013, 28, 1724–1727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santiago, J.A.; Potashkin, J.A. Evaluation of RNA Blood Biomarkers in Individuals at Risk of Parkinson’s Disease. J. Park. Dis. 2017, 7, 653–660. [Google Scholar] [CrossRef]
- Chikina, M.D.; Gerald, C.P.; Li, X.; Ge, Y.; Pincas, H.; Nair, V.D.; Wong, A.K.; Krishnan, A.; Troyanskaya, O.G.; Raymond, D.; et al. Low-variance RNAs identify Parkinson’s disease molecular signature in blood. Mov. Disord. Off. J. Mov. Disord. Soc. 2015, 30, 813–821. [Google Scholar] [CrossRef] [Green Version]
- Grünblatt, E. Parkinson’s disease: Molecular risk factors. Park. Relat. Disord. 2012, 18 (Suppl. S1), S45–S48. [Google Scholar] [CrossRef]
- Alieva, A.; Shadrina, M.I.; Filatova, E.V.; Karabanov, A.V.; Illarioshkin, S.N.; Limborska, S.A.; Slominsky, P.A. Involvement of endocytosis and alternative splicing in the formation of the pathological process in the early stages of Parkinson’s disease. BioMed Res. Int. 2014, 2014, 718732. [Google Scholar] [CrossRef] [Green Version]
- Smith, K.R.; Penzes, P. Ankyrins: Roles in synaptic biology and pathology. Mol. Cell. Neurosci. 2018, 91, 131–139. [Google Scholar] [CrossRef]
- Langston, J.W.; Ballard, P.; Tetrud, J.W.; Irwin, I. Chronic Parkinsonism in humans due to a product of meperidine-analog synthesis. Science 1983, 219, 979–980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Przedborski, S.; Jackson-Lewis, V.; Naini, A.B.; Jakowec, M.; Petzinger, G.; Miller, R.; Akram, M. The parkinsonian toxin 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP): A technical review of its utility and safety. J. Neurochem. 2001, 76, 1265–1274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lehtonen, Š.; Sonninen, T.M.; Wojciechowski, S.; Goldsteins, G.; Koistinaho, J. Dysfunction of Cellular Proteostasis in Parkinson’s Disease. Front. Neurosci. 2019, 13, 457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frøyset, A.K.; Edson, A.J.; Gharbi, N.; Khan, E.A.; Dondorp, D.; Bai, Q.; Tiraboschi, E.; Suster, M.L.; Connolly, J.B.; Burton, E.A.; et al. Astroglial DJ-1 over-expression up-regulates proteins involved in redox regulation and is neuroprotective in vivo. Redox Biol. 2018, 16, 237–247. [Google Scholar] [CrossRef]
- Virmani, A.; Gaetani, F.; Binienda, Z.; Xu, A.; Duhart, H.; Ali, S.F. Role of mitochondrial dysfunction in neurotoxicity of MPP+: Partial protection of PC12 cells by acetyl-L-carnitine. Ann. N. Y. Acad. Sci. 2004, 1025, 267–273. [Google Scholar] [CrossRef] [PubMed]






| Category | Term | Gene Symbol | FDR |
|---|---|---|---|
| GOTERM_BP_FAT | GO:0015671~oxygen transport | HBZ, HBM, HBD, HBQ1, BPGM | 0.000195516 |
| GOTERM_BP_FAT | GO:0061515~myeloid cell development | FAM210B, DMTN, EPB42, PIP4K2A, SLC4A1, BPGM, LTF | 0.000195516 |
| GOTERM_BP_FAT | GO:0015669~gas transport | HBZ, HBM, HBD, HBQ1, BPGM | 0.000489144 |
| GOTERM_BP_FAT | GO:0030218~erythrocyte differentiation | FAM210B, DMTN, EPB42, SLC4A1, AHSP, BPGM, KLF1 | 0.001167109 |
| GOTERM_BP_FAT | GO:0042744~hydrogen peroxide catabolic process | HBZ, HBM, HBD, HBQ1, SNCA | 0.001167109 |
| GOTERM_BP_FAT | GO:0034101~erythrocyte homeostasis | FAM210B, DMTN, EPB42, SLC4A1, AHSP, BPGM, KLF1 | 0.001324464 |
| GOTERM_BP_FAT | GO:0030099~myeloid cell differentiation | FAM210B, DMTN, EPB42, FAXDC2, PIP4K2A, SLC4A1, AHSP, BPGM, KLF1, LTF | 0.001445306 |
| GOTERM_BP_FAT | GO:0048821~erythrocyte development | FAM210B, DMTN, EPB42, SLC4A1, BPGM | 0.001476546 |
| GOTERM_BP_FAT | GO:0002262~myeloid cell homeostasis | FAM210B, DMTN, EPB42, SLC4A1, AHSP, BPGM, KLF1 | 0.003097455 |
| GOTERM_BP_FAT | GO:0042743~hydrogen peroxide metabolic process | HBZ, HBM, HBD, HBQ1, SNCA | 0.004875849 |
| GOTERM_BP_FAT | GO:0030097~hemopoiesis | FAM210B, DMTN, EPB42, GLRX5, FAXDC2, PIP4K2A, SLC4A1, AHSP, BPGM, FBXO7, KLF1, LTF | 0.023463887 |
| GOTERM_BP_FAT | GO:0048534~hematopoietic or lymphoid organ development | FAM210B, DMTN, EPB42, GLRX5, FAXDC2, PIP4K2A, SLC4A1, AHSP, BPGM, FBXO7, KLF1, LTF | 0.031640479 |
| GOTERM_BP_FAT | GO:0002520~immune system development | FAM210B, DMTN, EPB42, GLRX5, FAXDC2, PIP4K2A, SLC4A1, AHSP, BPGM, FBXO7, KLF1, LTF | 0.048049424 |
| GOTERM_BP_FAT | GO:0048872~homeostasis of number of cells | FAM210B, DMTN, EPB42, SLC4A1, AHSP, BPGM, KLF1 | 0.048049424 |
| GOTERM_CC_FAT | GO:0005833~hemoglobin complex | HBZ, HBM, AHSP, HBD, HBQ1 | 2.25 × 10−7 |
| GOTERM_CC_FAT | GO:0031838~haptoglobin-hemoglobin complex | HBZ, HBM, HBD, HBQ1 | 0.000819832 |
| GOTERM_CC_FAT | GO:0030863~cortical cytoskeleton | MPP1, TMOD1, DMTN, GYPC, EPB42, SLC4A1 | 0.004003729 |
| GOTERM_CC_FAT | GO:0044445~cytosolic part | HBZ, HBM, GLRX5, AHSP, HBD, HBQ1 | 0.020118051 |
| GOTERM_CC_FAT | GO:0044448~cell cortex part | MPP1, TMOD1, DMTN, GYPC, EPB42, SLC4A1 | 0.021770885 |
| GOTERM_CC_FAT | GO:0005938~cell cortex | MPP1, TMOD1, DMTN, GYPC, EPB42, SLC4A1, SNCA | 0.037572121 |
| GOTERM_MF_FAT | GO:0005344~oxygen transporter activity | HBZ, HBM, HBD, HBQ1 | 8.04107 × 10−5 |
| GOTERM_MF_FAT | GO:0031720~haptoglobin binding | HBZ, HBM, HBD, HBQ1 | 0.000809657 |
| GOTERM_MF_FAT | GO:0019825~oxygen binding | HBZ, HBM, HBD, HBQ1 | 0.00139728 |
| GOTERM_MF_FAT | GO:0005506~iron ion binding | HBZ, FECH, FAXDC2, HBQ1, LTF, SNCA | 0.024589133 |
| GOTERM_MF_FAT | GO:0030492~hemoglobin binding | SLC4A1, AHSP, HBD | 0.028801975 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xue, J.; Li, F.; Dai, P. The Potential of ANK1 to Predict Parkinson’s Disease. Genes 2023, 14, 226. https://doi.org/10.3390/genes14010226
Xue J, Li F, Dai P. The Potential of ANK1 to Predict Parkinson’s Disease. Genes. 2023; 14(1):226. https://doi.org/10.3390/genes14010226
Chicago/Turabian StyleXue, Jinsong, Fan Li, and Peng Dai. 2023. "The Potential of ANK1 to Predict Parkinson’s Disease" Genes 14, no. 1: 226. https://doi.org/10.3390/genes14010226