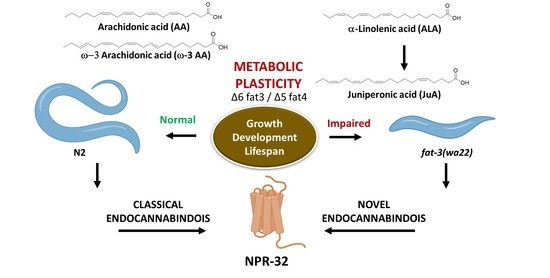
Juniperonic Acid Biosynthesis is Essential in Caenorhabditis elegans Lacking Δ6 Desaturase (fat-3) and Generates New ω-3 Endocannabinoids
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Growth Conditions
2.2. Dietary Lipid Supplementation
2.3. Brood Size Assay and Development Monitoring
2.4. Lifespan Assays
2.5. RNA Interference by Feeding
2.6. Synthesis of the Endocannabinoid-Like Molecules Juniperoyl Ethanolamide (JEA) and ω-3 Arachidonoyl Ethanolamine (ω-3 AEA)
2.7. Synthesis of the Endocannabinoid-Like Molecules 1/2-Juniperoylglycerol (1/2-JG) and ω-3 1/2-Arachidonoylglycerol (ω-3 1/2-AG)
2.8. Chromatographic Conditions Used for LC-MS/MS Targeted Metabolomics Analysis (Identification and Qquantification)
2.9. Quantitative Analysis of Targeted Analytes Using LC-MS/MS
2.10. Sample Preparation for LC-MS/MS Analysis
2.11. Biochemical Assays
2.12. NPR-32 Binding Assay
3. Results
3.1. Differential Expression of Eicosatetraenoic Acids in Wild Type and fat-3(wa22) Mutant C. elegans
3.2. Recovery of fat-3(wa22) Mutant Related Defects by Supplementation of JuA
3.3. Elucidation of JuA Biosynthesis in fat-3(wa22) Mutants
3.4. Detection of Different ω-3 Endocannabinoids in C. elegans WT and fat-3(wa22) Mutant Strain
3.5. Characterization of AA, ω-3 AA and JuA Aerived Endocannabinoids on Metabolic Enzymes and NPR-32 and NPR-19 Binding
4. Discussion
Author Contributions
Supplementary Materials
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| 1/2-AG | 1/2-arachidonoylglycerol |
| 1/2-JG | 1/2-juniperoylglycerol |
| 2-OG | 2-Mono-oleoyl-rac-glycerol |
| AA | Arachidonic acid |
| ACN | Acetonitrile |
| AEA | N-arachidonoyl ethanolamine |
| ALA | α-linoleic acid |
| CCR2 | C-C motif chemokine receptor 2 |
| CGC | Caenorhabditis Genetics Center |
| CHCl3 | Chloroform |
| CH3OH | Methanol |
| DAGL | Diacylglycerol lipase |
| DHGLA | Dihomo-gamma-linolenic acid |
| ECs | Endocannabinoids |
| ECS | Endocannabinoid system |
| ELO 1/2 | Fatty acid eongases in C. elegans |
| EPA | Eicosapentanenoic acid |
| FAs | Fatty acids |
| FAAH | Fatty acid amide hydrolase |
| FADS2 | Fatty acid desaturase 2, gene of Δ6 desaturase |
| FUDR | 5′-fluorodeoxyuridine |
| GLA | Gamma linoleic acid |
| GPCR | G protein coupled receptor |
| HCl | Hydrogen chloride |
| IPTG | Isopropyl-β-D-thiogalactopyranoside |
| JEA | N-juniperoyl ethanolamine |
| JuA | Juniperonic acid |
| LA | Linoleic acid |
| LC | Liquid chromatography |
| MAGL | Monoacylglycerol lipase |
| MgCl2 | Magnesium chloride |
| NAE | N-acylethanolamine |
| NAPE-PLD | N-acylphosphatidylethanolamine-hydrolyzing phospholipase D |
| NGM | Nematode growth media |
| NPR-19 | Cannabinoid G-protein coupled receptor in C. elegans |
| NPR-32 | Cannabinoid G-protein coupled receptor in C. elegans |
| PUFAs | Polyunsaturated fatty acids |
| ScA | Sciadonic acid |
| ω -3 AEA | Omega-3 N-arachidonoyl ethanolamine |
| ω -3 1/2-AG | Omega-3 1/2-arachidonoyl glycerol |
| ω-3 AA | Omega-3 arachidonic acid |
References
- Guillou, H.; Zadravec, D.; Martin, P.G.P.; Jacobsson, A. The Key Roles of Elongases and Desaturases in Mammalian Fatty Acid Metabolism: Insights from Transgenic Mice. Prog. Lipid Res. 2010, 49, 186–199. [Google Scholar] [CrossRef] [PubMed]
- Casares, D.; Escribá, P.V.; Rosselló, C.A. Membrane Lipid Composition: Effect on Membrane and Organelle Structure, Function and Compartmentalization and Therapeutic Avenues. Int. J. Mol. Sci. 2019, 20, 2167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tallima, H.; El Ridi, R. Arachidonic Acid: Physiological Roles and Potential Health Benefits—A Review. J. Adv. Res. 2018, 11, 33–41. [Google Scholar] [CrossRef]
- Massey, K.; Nicolaou, A. Lipidomics of Polyunsaturated-Fatty-Acid-Derived Oxygenated Metabolites. Biochem. Soc. Trans. 2011, 39, 1240–1246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freitas, H.; Isaac, A.; Malcher-Lopes, R.; Diaz, B.; Trevenzoli, I.; De Melo Reis, R. Polyunsaturated Fatty Acids and Endocannabinoids in Health and Disease. Nutr. Neurosci. 2017, 21, 695–714. [Google Scholar] [CrossRef]
- Gachet, M.; Schubert, A.; Calarco, S.; Boccard, J.; Gertsch, J. Targeted Metabolomics Shows Plasticity in the Evolution of Signaling Lipids and Uncovers Old and New Endocannabinoids in the Plant Kingdom. Sci. Rep. 2017, 7, 1–15. [Google Scholar] [CrossRef]
- Carlier, H.; Bernard, A.; Caselli, C. Digestion and Absorption of Polyunsaturated Fatty Acids. Reprod. Nutr. Dev. EDP Sci. 1991, 31, 475–500. [Google Scholar] [CrossRef]
- Rivers, J.; Sinclair, A.; Crawford, M. Inability of the Cat to Desaturate Essential Fatty Acids. Nature 1975, 258, 171–173. [Google Scholar] [CrossRef]
- Rivers, J.P.; Hassam, A.G.; Alderson, C. The Absence of Delta 6-Desaturase Activity in the Cat [Proceedings]. Proc. Nutr. Soc. 1976, 35, 67A–68A. [Google Scholar]
- Cho, H.; Nakamura, M.; Clarke, S. Cloning, Expression, and Nutritional Regulation of the Mammalian Δ-6 Desaturase. J. Biol. Chem. 1999, 274, 471–477. [Google Scholar] [CrossRef] [Green Version]
- Stoffel, W.; Holz, B.; Jenke, B.; Binczek, E.; Günter, R.H.; Kiss, C.; Karakesisoglou, I.; Thevis, M.; Weber, A.A.; Arnhold, S.; et al. Δ6-Desaturase (FADS2) Deficiency Unveils the Role of Omega3- and Omega6-Polyunsaturated Fatty Acids. EMBO J. 2008, 27, 2281–2292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stroud, C.K.; Nara, T.Y.; Manuel, Roqueta-Rivera; Radlowski, E.C.; Lawrence, P.; Zhang, Y.; Cho, B.H.; Segre, M.; Hess, R.A.; Brenna, J.T.; et al. Disruption of FADS2 Gene in Mice Impairs Male Reproduction and Causes Dermal and Intestinal Ulceration. J. Lipid Res. 2009, 50, 1870–1880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stoffel, W.; Hammels, I.; Jenke, B.; Binczek, E.; Schmidt-Soltau, I.; Brodesser, S.; Odenthal, M.; Thevis, M. Obesity Resistance and Deregulation of Lipogenesis in Δ6-fatty Acid Desaturase (FADS 2) Deficiency. EMBO Rep. 2014, 15, 110–120. [Google Scholar] [CrossRef] [PubMed]
- Williard, D.; Nwankwo, J.; Kaduce, T.; Harmon, S.; Irons, M.; Moser, H.; Raymond, G.; Spector, A. Identification of a Fatty Acid Delta6-Desaturase Deficiency in Human Skin Fibroblasts. J. Lipid Res. 2001, 42, 501–508. [Google Scholar] [PubMed]
- Horrobin, D.F. Loss of Delta-6-Desaturase Activity as a Key Factor in Aging. Med. Hypotheses 1981, 7, 1211–1220. [Google Scholar] [CrossRef]
- Guarente, L.; Kenyon, C. Genetic Pathways That Regulate Ageing in Model Organisms. Nature 2000, 408, 255–262. [Google Scholar] [CrossRef]
- Kim, D.; Feinbaum, R.; Alloing, G.; Emerson, F.; Garsin, D.; Inoue, H.; Tanaka-Hino, M.; Hisamoto, N.; Matsumoto, K.; Tan, M.; et al. A Conserved P38 MAP Kinase Pathway in Caenorhabditis Elegans Innate Immunity. Science 2002, 297, 623–626. [Google Scholar] [CrossRef] [Green Version]
- Lee, R.; Hench, J.; Ruvkun, G. Regulation of C. Elegans DAF-16 and Its Human Ortholog FKHRL1 by the Daf-2 Insulin-like Signaling Pathway. Curr. Biol. 2001, 11, 1950–1957. [Google Scholar] [CrossRef] [Green Version]
- Baumeister, R.; Schaffitzel, E.; Hertweck, M. Endocrine Signaling in Caenorhabditis Elegans Controls Stress Response and Longevity. J. Endocrinol. 2006, 190, 191–202. [Google Scholar] [CrossRef]
- Yuan, J. Evolutionary Conservation of a Genetic Pathway of Programmed Cell Death. J. Cell Biochem. 1996, 60, 4–11. [Google Scholar] [CrossRef]
- Watts, J. Fat Synthesis and Adiposity Regulation in Caenorhabditis Elegans. Trends Endocrinol. Metab. 2009, 20, 58–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watts, J.L.; Browse, J. Genetic Dissection of Polyunsaturated Fatty Acid Synthesis in Caenorhabditis Elegans. Proc. Natl. Acad. Sci. USA 2002, 99, 5854–5859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spychalla, J.P.; Kinney, A.J.; Browse, J. Identification of an Animal Omega-3 Fatty Acid Desaturase by Heterologous Expression in Arabidopsis. PNAS 1997, 94, 1142–1147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peyou-Ndi, M.; Watts, J.; Browse, J. Identification and Characterization of an Animal Delta(12) Fatty Acid Desaturase Gene by Heterologous Expression in Saccharomyces Cerevisiae. Arch. Biochem. Biophys. 2000, 376, 399–408. [Google Scholar] [CrossRef] [PubMed]
- Lesa, G.M.; Palfreyman, M.; Hall, D.H.; Clandinin, M.T.; Rudolph, C.; Jorgensen, E.M.; Schiavo, G. Long Chain Polyunsaturated Fatty Acids Are Required for Efficient Neurotransmission in C. Elegans. J. Cell Sci. 2003, 116, 4965–4975. [Google Scholar] [CrossRef] [Green Version]
- Los, D.A.; Murata, N. Structure and Expression of Fatty Acid Desaturases. Biochim. Biophys. Acta–Lipids Lipid Metab. 1998, 1394, 3–15. [Google Scholar] [CrossRef]
- Napier, J.A.; Hey, S.J.; Lacey, D.J.; Shewry, P.R. Identification of a Caenorhabditis Elegans Delta6-Fatty-Acid-Desaturase by Heterologous Expression in Saccharomyces Cerevisiae. Biochem. J. 1998, 330, 611–614. [Google Scholar] [CrossRef]
- Napier, J.A.; Michaelson, L.V.; Stobart, A.K. Plant Desaturases: Harvesting the Fat of the Land. Curr. Opin. Plant Biol. 1999, 2, 123–127. [Google Scholar] [CrossRef]
- Kahn-Kirby, A.H.; Dantzker, J.L.M.; Apicella, A.J.; Schafer, W.R.; Browse, J.; Bargmann, C.I.; Watts, J.L. Specific Polyunsaturated Fatty Acids Drive TRPV-Dependent Sensory Signaling in Vivo. Cell 2004, 119, 889–900. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Falck, J.R.; Rothe, M.; Schunck, W.H.; Menzel, R. Role of CYP Eicosanoids in the Regulation of Pharyngeal Pumping and Food Uptake in Caenorhabditis Elegans. J. Lipid Res. 2015, 56, 2110–2123. [Google Scholar] [CrossRef] [Green Version]
- Pastuhov, S.I.; Matsumoto, K.; Hisamoto, N. Endocannabinoid Signaling Regulates Regenerative Axon Navigation in Caenorhabditis Elegans via the GPCRs NPR-19 and NPR-32. Genes Cells 2016, 21, 696–705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaudhuri, J.; Parihar, M.; Pires-daSilva, A. An Introduction to Worm Lab: From Culturing Animals to Mutagenesis. J. Vis. Exp. 2011, 47, 5–8. [Google Scholar]
- Ahringer, J. Reverse genetics, WormBook. The C. elegans Research Community; WormBook: Pasadena, CA, USA, 2006; pp. 1–43. [Google Scholar] [CrossRef]
- Hillyard, S.; German, J. Quantitative Lipid Analysis and Life Span of the Fat-3 Mutant of Caenorhabditis Elegans. Hillyard SL1, German JB. J. Agric. Food Chem. 2009, 57, 3389–3396. [Google Scholar] [CrossRef] [PubMed]
- Reisner, K.; Lehtonen, M.; Storvik, M.; Jantson, T.; Lakso, M.; Callaway, J.; Wong, G. Trans Fat Diet Causes Decreased Brood Size and Shortened Lifespan in Caenorhabditis Elegans Delta-6-Desaturase Mutant Fat-3. J. Biochem. Mol. Toxicol. 2011, 25, 269–279. [Google Scholar] [CrossRef]
- Watts, J.L.; Phillips, E.; Griffing, K.R.; Browse, J. Deficiencies in C20 Polyunsaturated Fatty Acids Cause Behavioral and Developmental Defects in Caenorhabditis Elegans Fat-3 Mutants. Genetics 2003, 163, 581–589. [Google Scholar]
- Watts, J.L. Using Caenorhabditis Elegans to Uncover Conserved Functions of Omega-3 and Omega-6 Fatty Acids. J. Clin. Med. 2016, 5, 19. [Google Scholar] [CrossRef] [Green Version]
- Kniazeva, M.; Sieber, M.; McCauley, S.; Zhang, K.; Watts, J.L.; Han, M. Suppression of the ELO-2 FA Elongation Activity Results in Alterations of the Fatty Acid Composition and Multiple Physiological Defects, Including Abnormal Ultradian Rhythms, in Caenorhabditis Elegans. Genetics 2003, 163, 159–169. [Google Scholar]
- Gachet, M.; Rhyn, P.; Bosch, O.; Quednow, B.B.; Gertsch, J. A Quantitiative LC-MS/MS Method for the Measurement of Arachidonic Acid, Prostanoids, Endocannabinoids, N-Acylethanolamines and Steroids in Human Plasma. J. Chromatogr. B. Anal. Technol. Biomed. Life Sci. 2015, 976, 6–18. [Google Scholar] [CrossRef]
- Fowler, C.J.; Jonsson, K.O.; Tiger, G. Fatty Acid Amide Hydrolase: Biochemistry, Pharmacology, and Therapeutic Possibilities for an Enzyme Hydrolyzing Anandamide, 2-Arachidonoylglycerol, Palmitoylethanolamide, and Oleamide. Biochem. Pharmacol. 2001, 62, 517–526. [Google Scholar] [CrossRef]
- Ullman, D.; Sprecher, H. An in Vitro and in Vivo Study of the Conversion of Eicosa-11, 14-Dienoic Acid to Eicosa-5,11,14-Trienoic Acid and of the Conversion of Eicosa-11-Enoic Acid to Eicosa-5,11-Dienoic Acid in the Rat. Biochim. Biophys. Acta. (BBA)—Lipids Lipid Metab. 1971, 248, 186–197. [Google Scholar] [CrossRef]
- Pertwee, R.G. Pharmacology of Cannabinoid Receptor Ligands. Curr. Med. Chem. 1999, 6, 635–664. [Google Scholar] [PubMed]
- Kapur, A.; Zhao, P.; Sharir, H.; Bai, Y.; Caron, M.G.; Barak, L.S.; Abood, M.E. Atypical Responsiveness of the Orphan Receptor GPR55 to Cannabinoid Ligands. J. Biol. Chem. 2009, 284, 29817–29827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bentsen, H. Dietary Polyunsaturated Fatty Acids, Brain Function and Mental Health. Microb. Ecol. Health Dis. 2017, 28, 1281916. [Google Scholar] [CrossRef]
- Rapoport, S.I. Arachidonic Acid and the Brain. J. Nutr. 2008, 138, 2515–2520. [Google Scholar] [CrossRef] [PubMed]
- Monk, J.M.; Liddle, D.M.; Cohen, D.J.A.; Tsang, D.H.; Hillyer, L.M.; Abdelmagid, S.A.; Nakamura, M.T.; Power, K.A.; Ma, D.W.L.; Robinson, L.E. The Delta 6 Desaturase Knock out Mouse Reveals That Immunomodulatory Effects of Essential N-6 and n-3 Polyunsaturated Fatty Acids Are Both Independent of and Dependent upon Conversion. J. Nutr. Biochem. 2016, 32, 29–38. [Google Scholar] [CrossRef] [Green Version]
- Dinh, T.K.; Bourre, J.M.; Durand, G. Effect of Age and Alpha-Linolenic Acid Deficiency on Delta 6 Desaturase Activity and Liver Lipids in Rats. Lipids 1993, 28, 517–523. [Google Scholar] [CrossRef]
- Sergeant, S.; Rahbar, E.; Chilton, F.H. Gamma-Linolenic Acid, Dihommo-Gamma Linolenic, Eicosanoids and Inflammatory Processes. Eur. J. Pharmacol. 2016, 785, 77–86. [Google Scholar] [CrossRef] [Green Version]
- Hammels, I.; Binczek, E.; Schmidt-Soltau, I.; Jenke, B.; Thomas, A.; Vogel, M.; Thevis, M.; Filipova, D.; Papadopoulos, S.; Stoffel, W. Novel CB1-Ligands Maintain Homeostasis of the Endocannabinoid System in Ω3- and Ω6-Long-Chain-PUFA Deficiency. J. Lipid Res. 2019, 60, 1396–1409. [Google Scholar] [CrossRef] [Green Version]
- Fan, Y.Y.; Chapkin, R.S. Importance of Dietary γ-Linolenic Acid in Human Health and Nutrition. J. Nutr. 1998, 128, 1411–1414. [Google Scholar] [CrossRef] [Green Version]
- Brenna, J.T. Long-Chain Polyunsaturated Fatty Acids and the Preterm Infant: A Case Study in Developmentally Sensitive Nutrient Needs in the United States. Am. J. Clin. Nutr. 2016, 103, 606S–615S. [Google Scholar] [CrossRef] [Green Version]
- Schlenk, H.; Gellerman, J.L. Arachidonic, 5, 11, 14, 17-eicosatetraenoic and Related Acids in Plants Identification of Unsaturated Fatty Acids. JAOCS 1965, 42, 504–511. [Google Scholar] [CrossRef]
- Ikeda, I.; Oka, T.; Koba, K.; Sugano, M.; Jie, M.S.F.L.K. 5c, 11c, 14c-Eicosatrienoic Acid and 5c, 11c, 14c, 17c-eicosatetraenoic Acid OfBiota Orientalis Seed Oil Affect Lipid Metabolism in the Rat. Lipids 1992, 27, 500–504. [Google Scholar] [CrossRef] [PubMed]
- Anderson, E.; Corkins, M.; Li, J.; Singh, K.; Parsons, S.; Tucey, T.; Sorkaç, A.; Huang, H.; Dimitriadi, M.; Sinclair, D.; et al. Elegans Lifespan Extension by Osmotic Stress Requires FUdR, Base Excision Repair, FOXO, and Sirtuins. Mech. Ageing Dev. 2016, 154, 30–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kato, Y.; Miyaji, M.; Zhang-Akiyama, Q. FUdR Extends the Lifespan of the Short-Lived AP Endonuclease Mutant in Caenorhabditis Elegans in a Fertility-Dependent Manner. Genes Genet. Syst. 2017, 91, 201–207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Raamsdonk, J.; Hekimi, S. FUdR Causes a Twofold Increase in the Lifespan of the Mitochondrial Mutant Gas-1. Mech. Ageing Dev. 2011, 132, 519–521. [Google Scholar] [CrossRef] [Green Version]
- Aitlhadj, L.; Stürzenbaum, S. The Use of FUdR Can Cause Prolonged Longevity in Mutant Nematodes. Mech. Ageing Dev. 2010, 131, 364–365. [Google Scholar] [CrossRef]
- Vásquez, V.; Krieg, M.; Lockhead, D.; Goodman, M.B. Phospholipids That Contain Polyunsaturated Fatty Acids Enhance Neuronal Cell Mechanics and Touch Sensation. Cell Rep. 2014, 6, 70–80. [Google Scholar] [CrossRef] [Green Version]
- Lauretani, F.; Bandinelli, S.; Benedetta, B.; Cherubini, A.; Iorio, A.D.; Blè, A.; Giacomini, V.; Corsi, A.M.; Guralnik, J.M.; Ferrucci, L. Omega-6 and Omega-3 Fatty Acids Predict Accelerated Decline of Peripheral Nerve Function in Older Persons. Eur. J. Neurol. 2007, 14, 801–808. [Google Scholar] [CrossRef] [Green Version]
- Tsai, P.J.; Huang, W.C.; Lin, S.W.; Chen, S.N.; Shen, H.J.; Chang, H.; Chuang, L.T. Juniperonic Acid Incorporation into the Phospholipids of Murine Macrophage Cells Modulates Pro-Inflammatory Mediator Production. Inflammation 2018, 41, 1200–1214. [Google Scholar] [CrossRef]
- Tanaka, T.; Morishige, J.; Iwawaki, D.; Fukuhara, T.; Hamamura, N.; Hirano, K.; Osumi, T.; Satouchi, K. Metabolic Pathway That Produces Essential Fatty Acids from Polymethylene-Interrupted Polyunsaturated Fatty Acids in Animal Cells. FEBS J. 2007, 274, 2728–2737. [Google Scholar] [CrossRef]
- WormBase Nematode Information Resource Website Version: WS277. Available online: http://www.wormbase.org/db/get?name=WBGene00001240,class=Gene (accessed on 17 September 2012).
- Watts, J.L.; Browse, J. Isolation and Characterization of a ∆5-Fatty Acid Desaturase from Caenorhabditis Elegans. Arch. Biochem. Biophys. 1999, 362, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.C.; Mackie, K. An Introduction to the Endogenous Cannabinoid System Hui-Chen. Biol. Psychiatry. 2016, 79, 516–525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harrison, N.; Lone, M.A.; Kaul, T.K.; Rodrigues, P.R.; Ogungbe, I.V.; Gill, M.S. Characterization of N-Acyl Phosphatidylethanolamine-Specific Phospholipase-D Isoforms in the Nematode Caenorhabditis Elegans. PLoS One 2014, 9, e113007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galles, C.; Prez, G.M.; Penkov, S.; Boland, S.; Porta, E.O.J.; Altabe, S.G.; Labadie, G.R.; Schmidt, U.; Knölker, H.J.; Kurzchalia, T.V.; et al. Endocannabinoids in Caenorhabditis Elegans Are Essential for the Mobilization of Cholesterol from Internal Reserves. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef]
- Chen, A.L.; Lum, K.M.; Lara-Gonzalez, P.; Ogasawara, D.; Cognetta, A.B.; To, A.; Parsons, W.H.; Simon, G.M.; Desai, A.; Petrascheck, M.; et al. Pharmacological Convergence Reveals a Lipid Pathway That Regulates, C. Elegans Lifespan. Nat. Chem. Biol. 2019, 15, 453–462. [Google Scholar] [CrossRef]
- Lucanic, M.; Held, J.M.; Vantipalli, M.C.; Klang, I.M.; Graham, J.B.; Gibson, B.W.; Lithgow, G.J.; Gill, M.S. N-Acylethanolamine Signalling Mediates the Effect of Diet on Lifespan in Caenorhabditis Elegans. Nature 2011, 473, 226–229. [Google Scholar] [CrossRef]
- Kohno, M.; Hasegawa, H.; Inoue, A.; Muraoka, M.; Miyazaki, T.; Oka, K.; Yasukawa, M. Identification of N-Arachidonylglycine as the Endogenous Ligand for Orphan G-Protein-Coupled Receptor GPR18. Biochem. Biophys. Res. Commun. 2006, 347, 827–832. [Google Scholar] [CrossRef]
- Johns, D.G.; Behm, D.J.; Walker, D.J.; Ao, Z.; Shapland, E.M.; Daniels, D.A.; Riddick, M.; Dowell, S.; Staton, P.C.; Green, P.; et al. The Novel Endocannabinoid Receptor GPR55 Is Activated by Atypical Cannabinoids but Does Not Mediate Their Vasodilator Effects. Br. J. Pharmacol. 2007, 152, 825–831. [Google Scholar] [CrossRef] [Green Version]
- Knerlich-Lukoschus, F.; Noack, M.; von der Ropp-Brenner, B.; Lucius, R.; Mehdorn, H.M.; Held-Feindt, J. Spinal Cord Injuries Induce Changes in CB1 Cannabinoid Receptor and C-C Chemokine Expression in Brain Areas Underlying Circuitry of Chronic Pain Conditions. J. Neurotrauma 2011, 28, 619–634. [Google Scholar] [CrossRef]
- Oakes, M.D.; Law, W.J.; Clark, T.; Bamber, B.A.; Komuniecki, R. Cannabinoids Activate Monoaminergic Signaling to Modulate Key, C. elegans Behaviors. J. Neurosci. 2017, 37, 2859–2869. [Google Scholar] [CrossRef] [Green Version]
- Bilkei-Gorzo, A. The Endocannabinoid System in Normal and Pathological Brain Ageing. Philos. Trans R. Soc. L. B. Biol. Sci. 2012, 367, 3326–3341. [Google Scholar] [CrossRef] [PubMed]
- Watkins, B.A.; Kim, J. The Endocannabinoid System: Directing Eating Behavior and Macronutrient Metabolism. Front Psychol. 2014, 5, 1506. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, S.; Tam, J. Cannabis: From a Plant That Modulates Feeding Behaviors toward Developing Selective Inhibitors of the Peripheral Endocannabinoid System for the Treatment of Obesity and Metabolic Syndrome. Toxins 2019, 11, 275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García-Rincón, D.; Díaz-Alonso, J.; Paraíso-Luna, J.; Ortega, Z.; Aguareles, J.; de Salas-Quiroga, A.; Jou, C.; de Prada, I.; Martínez-Cerdeño, V.; Aronica, E.; et al. Contribution of Altered Endocannabinoid System to Overactive MTORC1 Signaling in Focal Cortical Dysplasia. Front. Pharmacol. 2018, 9, 1508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coccurello, R.; Maccarrone, M. Hedonic Eating and the “Delicious Circle”: From Lipid-Derived Mediators to Brain Dopamine and Back. Front Neurosci. 2018, 12, 271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tosi, F.; Sartori, F.; Guarini, P.; Olivieri, O.; Martinelli, N. Delta-5 and delta-6 desaturases: crucial enzymes in polyunsaturated fatty acid-related pathways with pleiotropic influences in health and disease. Adv. Clin. Exp. Med. 2014, 824, 61. [Google Scholar]










| Fatty Acids | WT | fat-3 (wa22) |
|---|---|---|
| Arachidonic acid (AA) | + | - |
| ω-3 arachidonic acid (ω-3 AA) | + | - |
| Juniperonic acid (JuA) | - | + |
| α-linoleic acid (ALA) | - | + |
| γ-linoleic acid (GLA) | + | - |
| Sciadonic acid (ScA) | - | - |
| dihomo-γ-linoleic acid (DHGLA) | + | - |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guha, S.; Calarco, S.; Gachet, M.S.; Gertsch, J. Juniperonic Acid Biosynthesis is Essential in Caenorhabditis elegans Lacking Δ6 Desaturase (fat-3) and Generates New ω-3 Endocannabinoids. Cells 2020, 9, 2127. https://doi.org/10.3390/cells9092127
Guha S, Calarco S, Gachet MS, Gertsch J. Juniperonic Acid Biosynthesis is Essential in Caenorhabditis elegans Lacking Δ6 Desaturase (fat-3) and Generates New ω-3 Endocannabinoids. Cells. 2020; 9(9):2127. https://doi.org/10.3390/cells9092127
Chicago/Turabian StyleGuha, Sujay, Serafina Calarco, M. Salomé Gachet, and Jürg Gertsch. 2020. "Juniperonic Acid Biosynthesis is Essential in Caenorhabditis elegans Lacking Δ6 Desaturase (fat-3) and Generates New ω-3 Endocannabinoids" Cells 9, no. 9: 2127. https://doi.org/10.3390/cells9092127