Qualifying Osteogenic Potency Assay Metrics for Human Multipotent Stromal Cells: TGF-β2 a Telling Eligible Biomarker
Abstract
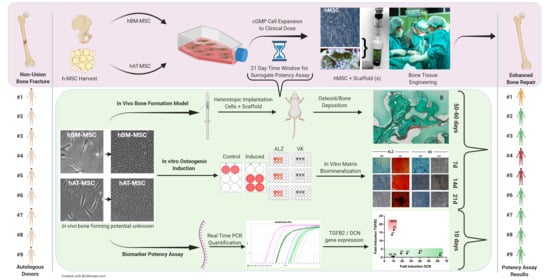
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Induction of Ex Vivo Osteogenic Differentiation
2.3. Matrix Mineralization Assays
2.4. RNA Extraction and Relative Quantification by Real-Time PCR
2.5. Median Absolute Dispersion (MAD) Analyses for Household Genes Selection
2.6. Hierarchical Cluster Analysis
2.7. In Vivo Heterotopic Bone Formation
2.8. Histology and Immunohistochemistry
3. Results
3.1. Establishment of hBM-MSC and hAT-MSC Expansion in Culture
3.2. Osteogenic Medium Promptly Induced hBM-MSC and hAT-MSC Collagen Matrix Biomineralization
3.3. The Real-Time PCR Reference Gene Pair of Lowest Median Absolute Dispersion (MAD) Was GUSB/YWAHZ
3.4. Inter-Donor Heterogeneity for Osteogenic Potency Signature Gene Induction in hBM-MSC and hAT-MSC
3.5. TaqMan Real-Time PCR Confirmed Low Induction of COL1A2, ELN and RUNX2 Genes
3.6. Cross-Laboratory Comparison of Potency Assay Cluster Analysis
3.7. TGFB2 Gene Induction Was Distinct for hBM-MSC Versus hAT-MSC
3.8. Donor-Dependent Bone Formation by hBM-MSC and hAT-MSC with Distinct Decorin and TGF-β2 Immunohistology
3.9. Inversely Correlated TGFB2 and DCN Gene Induction and Prospective Recognition of hMSC Bone-Forming Potential In Vivo
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lin, H.; Sohn, J.; Shen, H.; Langhans, M.T.; Tuan, R.S. Bone marrow mesenchymal stem cells: Aging and tissue engineering applications to enhance bone healing. Biomaterials 2019, 203, 96–110. [Google Scholar] [CrossRef] [PubMed]
- Quarto, R.; Mastrogiacomo, M.; Cancedda, R.; Kutepov, S.M.; Mukhachev, V.; Lavroukov, A.; Kon, E.; Marcacci, M. Repair of large bone defects with the use of autologous bone marrow stromal cells. N. Engl. J. Med. 2001, 344, 385–386. [Google Scholar] [CrossRef] [PubMed]
- Imam, M.A.; Holton, J.; Ernstbrunner, L.; Pepke, W.; Grubhofer, F.; Narvani, A.; Snow, M. A systematic review of the clinical applications and complications of bone marrow aspirate concentrate in management of bone defects and nonunions. Int. Orthop. 2017, 41, 2213–2220. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Barrena, E.; Padilla-Eguiluz, N.; Rosset, P.; Gebhard, F.; Hernigou, P.; Baldini, N.; Rouard, H.; Sensebé, L.; Gonzalo-Daganzo, R.-M.; Giordano, R.; et al. Early efficacy evaluation of mesenchymal stromal cells (MSC) combined to biomaterials to treat long bone non-unions. Injury 2020, 51 (Suppl. 1), S63–S73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siddappa, R.; Licht, R.; van Blitterswijk, C.; de Boer, J. Donor variation and loss of multipotency during in vitro expansion of human mesenchymal stem cells for bone tissue engineering. J. Orthop. Res. 2007, 25, 1029–1041. [Google Scholar] [CrossRef] [PubMed]
- Costa, L.A.; Eiro, N.; Fraile, M.; Gonzalez, L.O.; Saá, J.; Garcia-Portabella, P.; Vega, B.; Schneider, J.; Vizoso, F.J. Functional heterogeneity of mesenchymal stem cells from natural niches to culture conditions: Implications for further clinical uses. Cell Mol. Life Sci. 2020. [Google Scholar] [CrossRef]
- O’Keefe, R.J.; Tuan, R.S.; Lane, N.E.; Awad, H.; Barry, F.; Bunnell, B.A.; Colnot, C.; Drake, M.T.; Drissi, H.; A Dyment, N.; et al. American Society for Bone and Mineral Research-Orthopaedic Research Society Joint Task Force Report on Cell-Based Therapies. J. Bone Min. Res. 2020, 35, 3–17. [Google Scholar] [CrossRef]
- Pimpaneau, V.; Gianelli, F.; Trouvin, J.-H.; Poiseau, A.D. The challenges of potency assay development for cell-based medicinal products in Europe. Regul. Rapp. 2015, 12, 5–10. [Google Scholar]
- Gómez-Barrena, E.; Rosset, P.; Gebhard, F.; Hernigou, P.; Baldini, N.; Rouard, H.; Sensebé, L.; Gonzalo-Daganzo, R.M.; Giordano, R.; Padilla-Eguiluz, N.; et al. Feasibility and safety of treating non-unions in tibia, femur and humerus with autologous, expanded, bone marrow-derived mesenchymal stromal cells associated with biphasic calcium phosphate biomaterials in a multicentric, non-comparative trial. Biomaterials 2019, 196, 100–108. [Google Scholar] [CrossRef]
- Fiorentini, E.; Granchi, D.; Leonardi, E.; Baldini, N.; Ciapetti, G. Effects of Osteogenic Differentiation Inducers on in Vitro Expanded Adult Mesenchymal Stromal Cells. Int. J. Artif. Organs 2011, 34, 998–1011. [Google Scholar] [CrossRef]
- Lian, J.B.; Stein, G.S. Concepts of osteoblast growth and differentiation: Basis for modulation of bone cell development and tissue formation. Crit. Rev. Oral Biol. Med. 1992, 3, 269–305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rutkovskiy, A.; Stensløkken, K.O.; Vaage, I.J. Osteoblast Differentiation at a Glance. Med. Sci. Monit. Basic Res. 2016, 22, 95–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, B.; Menzel, U.; Loebel, C.; Schmal, H.; Alini, M.; Stoddart, M.J. Monitoring live human mesenchymal stromal cell differentiation and subsequent selection using fluorescent RNA-based probes. Sci. Rep. 2016, 6, 26014. [Google Scholar] [CrossRef] [Green Version]
- Lin, X.; Patil, S.; Gao, Y.-G.; Qian, A. The Bone Extracellular Matrix in Bone Formation and Regeneration. Front. Pharmacol. 2020, 11, 757. [Google Scholar] [CrossRef] [PubMed]
- Hoshiba, T.; Kawazoe, N.; Tateishi, T.; Chen, G. Development of stepwise osteogenesis-mimicking matrices for the regulation of mesenchymal stem cell functions. J. Biol. Chem. 2009, 284, 31164–31173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larsen, K.H.; Frederiksen, C.M.; Burns, J.S.; Abdallah, B.M.; Kassem, M. Identifying a molecular phenotype for bone marrow stromal cells with in vivo bone-forming capacity. J. Bone Min. Res. 2010, 25, 796–808. [Google Scholar]
- Zupan, J.; Tang, D.; Oreffo, R.O.C.; Redl, H.; Presen, D.M. Cell Engineering and Regeneration; Springer International Publishing: Berlin/Heidelberg, Germany, 2020; p. 139. [Google Scholar]
- Murgia, A.; Veronesi, E.; Candini, O.; Caselli, A.; D’Souza, N.; Rasini, V.; Giorgini, A.; Catani, F.; Iughetti, L.; Dominici, M.; et al. Potency Biomarker Signature Genes from Multiparametric Osteogenesis Assays: Will cGMP Human Bone Marrow Mesenchymal Stromal Cells Make Bone. PLoS ONE 2016, 11, e0163629. [Google Scholar] [CrossRef] [PubMed]
- Amărandi, R.M.; Becheru, D.F.; Vlăsceanu, G.M.; Ioniță, M.; Burns, J.S. Advantages of Graphene Biosensors for Human Stem Cell Therapy Potency Assays. Biomed. Res. Int. 2018, 2018, 1676851. [Google Scholar] [CrossRef]
- Halvorsen, Y.C.; Wilkison, W.O.; Gimble, J.M. Adipose-derived stromal cells—Their utility and potential in bone formation. Int. J. Obes. 2000, 24, S41–S44. [Google Scholar] [CrossRef] [Green Version]
- Shafiee, A.; Seyedjafari, E.; Soleimani, M.; Ahmadbeigi, N.; Dinarvand, P.; Ghaemi, N. A comparison between osteogenic differentiation of human unrestricted somatic stem cells and mesenchymal stem cells from bone marrow and adipose tissue. Biotechnol. Lett. 2011, 33, 1257–1264. [Google Scholar] [CrossRef]
- Strioga, M.; Viswanathan, S.; Darinskas, A.; Slaby, O.; Michalek, J. Same or not the same? Comparison of adipose tissue-derived versus bone marrow-derived mesenchymal stem and stromal cells. Stem Cells Dev. 2012, 21, 2724–2752. [Google Scholar] [CrossRef] [PubMed]
- Ciuffi, S.; Zonefrati, R.; Brandi, M.L. Adipose stem cells for bone tissue repair. Clin. Cases Miner. Bone Metab. 2017, 14, 217. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Goulart, V.; Ferreira, L.B.; Duarte, C.A.; de Lima, I.L.; Ferreira, E.R.; de Oliveira, B.C.; Vargas, L.N.; de Moraes, D.D.; Silva, I.B.B.; de Oliveira Faria, R. Mesenchymal stem cells from human adipose tissue and bone repair: A literature review. Biotechnol. Res. Innov. 2018, 2, 74–80. [Google Scholar] [CrossRef]
- Mohamed-Ahmed, S.; Fristad, I.; Lie, S.A.; Suliman, S.; Mustafa, K.; Vindenes, H.; Idris, S.B. Adipose-derived and bone marrow mesenchymal stem cells: A donor-matched comparison. Stem Cell Res. 2018, 9, 168. [Google Scholar] [CrossRef]
- González-Bermúdez, L.; Anglada, T.; Genescà, A.; Martín, M.; Terradas, M. Identification of reference genes for RT-qPCR data normalisation in aging studies. Sci. Rep. 2019, 9, 13970. [Google Scholar] [CrossRef] [Green Version]
- Ragni, E.; Viganò, M.; Rebulla, P.; Giordano, R.; Lazzari, L. What is beyond a qRT-PCR study on mesenchymal stem cell differentiation properties: How to choose the most reliable housekeeping genes. J. Cell Mol. Med. 2013, 17, 168–180. [Google Scholar] [CrossRef]
- Christy, B.A.; Herzig, M.C.; Delavan, C.; Cantu, C.; Salgado, C.; Bynum, J.A.; Cap, A.P. Human primary fibroblasts perform similarly to MSCs in assays used to evaluate MSC safety and potency. Transfusion 2019, 59, 1593–1600. [Google Scholar] [CrossRef] [Green Version]
- Okajcekova, T.; Strnadel, J.; Pokusa, M.; Zahumenska, R.; Janickova, M.; Halasova, E.; Skovierova, H. A Comparative In Vitro Analysis of the Osteogenic Potential of Human Dental Pulp Stem Cells Using Various Differentiation Conditions. Int. J. Mol. Sci. 2020, 21, 2280. [Google Scholar] [CrossRef] [Green Version]
- Poniatowski, Ł.A.; Wojdasiewicz, P.; Gasik, R.; Szukiewicz, D. Transforming growth factor Beta family: Insight into the role of growth factors in regulation of fracture healing biology and potential clinical applications. Mediat. Inflamm. 2015, 2015, 137823. [Google Scholar] [CrossRef] [Green Version]
- Wu, M.; Chen, G.; Li, Y.P. TGF-β and BMP signaling in osteoblast, skeletal development, and bone formation, homeostasis and disease. Bone Res. 2016, 4, 16009. [Google Scholar] [CrossRef]
- Elsafadi, M.; Manikandan, M.; Almalki, S.; Mobarak, M.; Atteya, M.; Iqbal, Z.; Hashmi, J.A.; Shaheen, S.; Alajez, N.M.; Alfayez, M.; et al. TGFβ1-Induced Differentiation of Human Bone Marrow-Derived MSCs Is Mediated by Changes to the Actin Cytoskeleton. Stem Cells Int. 2018, 2018, 6913594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brennan, M.Á.; Renaud, A.; Amiaud, J.; Rojewski, M.T.; Schrezenmeier, H.; Heymann, D.; Trichet, V.; Layrolle, P. Pre-clinical studies of bone regeneration with human bone marrow stromal cells and biphasic calcium phosphate. Stem Cell Res. Ther. 2014, 5, 114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuk, P.A.; Zhu, M.; Ashjian, P.; De Ugarte, D.A.; Huang, J.I.; Mizuno, H.; Alfonso, Z.C.; Fraser, J.K.; Benhaim, P.; Hedrick, M.H. Human adipose tissue is a source of multipotent stem cells. Mol. Biol. Cell 2002, 13, 4279–4295. [Google Scholar] [CrossRef] [PubMed]
- Puchtler, H.; Meloan, S.N.; Terry, M.S. On the history and mechanism of alizarin and alizarin red S stains for calcium. J. Histochem. Cytochem. 1969, 17, 110–124. [Google Scholar] [CrossRef]
- Meloan, S.N.; Puchtler, H. Chemical Mechanisms of Staining Methods: Von Kossa’s Technique: What von Kossa Really Wrote and a Modified Reaction for Selective Demonstration of Inorganic Phosphates. J. Histotechnol. 1985, 8, 11–13. [Google Scholar] [CrossRef]
- Chomczynski, P. A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. Biotechniques 1993, 15, 532–534; 536–537. [Google Scholar]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper--Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome. Biol. 2002, 3, RESEARCH0034. [Google Scholar] [CrossRef] [Green Version]
- Abdallah, B.M.; Ditzel, N.; Kassem, M. Assessment of bone formation capacity using in vivo transplantation assays: Procedure and tissue analysis. Methods Mol. Biol. 2008, 455, 89–100. [Google Scholar]
- Burrow, K.L.; Hoyland, J.A.; Richardson, S.M. Human adipose-derived stem cells exhibit enhanced proliferative capacity and retain multipotency longer than donor-matched bone marrow mesenchymal stem cells during expansion in vitro. Stem Cells Int. 2017, 2017, 1–15. [Google Scholar] [CrossRef]
- Amable, P.R.; Teixeira, M.V.; Carias, R.B.; Granjeiro, J.M.; Borojevic, R. Identification of appropriate reference genes for human mesenchymal cells during expansion and differentiation. PLoS ONE 2013, 8, e73792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tajadini, M.; Panjehpour, M.; Javanmard, S.H. Comparison of SYBR Green and TaqMan methods in quantitative real-time polymerase chain reaction analysis of four adenosine receptor subtypes. Adv. Biomed. Res. 2014, 3, 85. [Google Scholar] [CrossRef] [PubMed]
- Maruelli, S.; Besio, R.; Rousseau, J.; Garibaldi, N.; Amiaud, J.; Brulin, B.; Layrolle, P.; Escriou, V.; Rossi, A.; Trichet, V.; et al. Osteoblasts mineralization and collagen matrix are conserved upon specific Col1a2 silencing. Matrix Biol. Plus 2020, 6–7, 100028. [Google Scholar] [CrossRef]
- Karamboulas, K.; Dranse, H.J.; Underhill, T.M. Regulation of BMP-dependent chondrogenesis in early limb mesenchyme by TGFbeta signals. J. Cell Sci. 2010, 123, 2068–2076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patrikoski, M.; Mannerström, B.; Miettinen, S. Perspectives for Clinical Translation of Adipose Stromal/Stem Cells. Stem Cells Int. 2019, 2019, 5858247. [Google Scholar] [CrossRef] [Green Version]
- Ntege, E.H.; Sunami, H.; Shimizu, Y. Advances in regenerative therapy: A review of the literature and future directions. Regen. Ther. 2020, 14, 136–153. [Google Scholar] [CrossRef]
- Liu, S.; De Castro, L.F.; Jin, P.; Civini, S.; Ren, J.; Reems, J.-A.; Cancelas, J.; Nayak, R.; Shaw, G.; O’Brien, T.; et al. Manufacturing Differences Affect Human Bone Marrow Stromal Cell Characteristics and Function: Comparison of Production Methods and Products from Multiple Centers. Sci. Rep. 2017, 7, 46731. [Google Scholar] [CrossRef]
- Phinney, D.G.; Kopen, G.; Righter, W.; Webster, S.; Tremain, N.; Prockop, D.J. Donor variation in the growth properties and osteogenic potential of human marrow stromal cells. J. Cell Biochem. 1999, 75, 424–436. [Google Scholar] [CrossRef]
- Ambrosi, T.H.; Longaker, M.T.; Chan, C.K.F. A Revised Perspective of Skeletal Stem Cell Biology. Front. Cell Dev. Biol. 2019, 7, 189. [Google Scholar] [CrossRef] [Green Version]
- Mastrolia, I.; Foppiani, E.M.; Murgia, A.; Candini, O.; Samarelli, A.V.; Grisendi, G.; Veronesi, E.; Horwitz, E.M.; Dominici, M. Challenges in Clinical Development of Mesenchymal Stromal/Stem Cells: Concise Review. Stem Cells Transl. Med. 2019, 8, 1135–1148. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, T.; Nakamura-Takahashi, A.; Kasahara, M.; Yamaguchi, A.; Azuma, T. Tissue-nonspecific alkaline phosphatase promotes the osteogenic differentiation of osteoprogenitor cells. Biochem. Biophys. Res. Commun. 2020, 524, 702–709. [Google Scholar] [CrossRef] [PubMed]
- Vimalraj, S. Alkaline phosphatase: Structure, expression and its function in bone mineralization. Gene 2020, 754, 144855. [Google Scholar] [CrossRef] [PubMed]
- Hempel, U.; Müller, K.; Preissler, C.; Noack, C.; Boxberger, S.; Dieter, P.; Bornhäuser, M.; Wobus, M. Human Bone Marrow Stromal Cells: A Reliable, Challenging Tool for In Vitro Osteogenesis and Bone Tissue Engineering Approaches. Stem Cells Int. 2016, 2016, 7842191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Augusciak-Duma, A.; Witecka, J.; Sieron, A.L.; Janeczko, M.; Pietrzyk, J.J.; Ochman, K.; Galicka, A.; Borszewska-Kornacka, M.K.; Pilch, J.; Jakubowska-Pietkiewicz, E. Mutations in the COL1A1 and COL1A2 genes associated with osteogenesis imperfecta (OI) types I or III. Acta Biochim. Pol. 2018, 65, 79–86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gorski, J.P.; Hankenson, K.D. Principles of Bone Biology; Elsevier: Amsterdam, The Netherlands, 2020; p. 359. [Google Scholar]
- Hoshi, K.; Kemmotsu, S.; Takeuchi, Y.; Amizuka, N.; Ozawa, H. The primary calcification in bones follows removal of decorin and fusion of collagen fibrils. J. Bone Min. Res. 1999, 14, 273–280. [Google Scholar] [CrossRef]
- Horiguchi, M.; Ota, M.; Rifkin, D.B. Matrix control of transforming growth factor-β function. J. Biochem. 2012, 152, 321–329. [Google Scholar] [CrossRef] [Green Version]
- Kuçi, S.; Kuçi, Z.; Schäfer, R.; Spohn, G.; Winter, S.; Schwab, M.; Salzmann-Manrique, E.; Klingebiel, T.; Bader, P. Molecular signature of human bone marrow-derived mesenchymal stromal cell subsets. Sci. Rep. 2019, 9, 1774. [Google Scholar] [CrossRef] [Green Version]
- Thiagarajan, L.; Abu-Awwad, H.A.-D.M.; Dixon, J.E. Osteogenic Programming of Human Mesenchymal Stem Cells with Highly Efficient Intracellular Delivery of RUNX2. Stem Cells Transl. Med. 2017, 6, 2146–2159. [Google Scholar] [CrossRef]
- Komori, T. Regulation of Proliferation, Differentiation and Functions of Osteoblasts by Runx2. Int. J. Mol. Sci. 2019, 20, 1694. [Google Scholar] [CrossRef] [Green Version]
- Boxall, S.A.; Jones, E. Markers for characterization of bone marrow multipotential stromal cells. Stem Cells Int. 2012, 2012, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Antebi, B.; Asher, A.M.; Rodriguez, L.A.; Moore, R.K.; Mohammadipoor, A.; Cancio, L.C. Cryopreserved mesenchymal stem cells regain functional potency following a 24-h acclimation period. J. Transl. Med. 2019, 17, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chevallier, N.; Anagnostou, F.; Zilber, S.; Bodivit, G.; Maurin, S.; Barrault, A.; Bierling, P.; Hernigou, P.; Layrolle, P.; Rouard, H. Osteoblastic differentiation of human mesenchymal stem cells with platelet lysate. Biomaterials 2010, 31, 270–278. [Google Scholar] [CrossRef] [PubMed]
- Shansky, Y.D.; Sergeeva, N.S.; Sviridova, I.K.; Karalkin, P.A.; Kirsanova, V.A.; Akhmedova, S.A.; Fomicheva, K.A.; Shkurnikov, M.Y.; Portyannikova, A.Y.; Kaprin, A.D. Human Platelet Lysate Sustains the Osteogenic/Adipogenic Differentiation Potential of Adipose-Derived Mesenchymal Stromal Cells and Maintains Their DNA Integrity in vitro. Cells Tissues Organs 2019, 207, 149–164. [Google Scholar] [CrossRef] [PubMed]
- Turinetto, V.; Vitale, E.; Giachino, C. Senescence in Human Mesenchymal Stem Cells: Functional Changes and Implications in Stem Cell-Based Therapy. Int. J. Mol. Sci. 2016, 17, 1164. [Google Scholar] [CrossRef]
- Li, X.; Yang, Q.; Bai, J.; Xuan, Y.; Wang, Y. Identification of appropriate reference genes for human mesenchymal stem cell analysis by quantitative real-time PCR. Biotechnol. Lett. 2015, 37, 67–73. [Google Scholar] [CrossRef] [Green Version]
- Burns, J.S.; Rasmussen, P.L.; Larsen, K.H.; Schroder, H.D.; Kassem, M. Parameters in three-dimensional osteospheroids of telomerized human mesenchymal (stromal) stem cells grown on osteoconductive scaffolds that predict in vivo bone-forming potential. Tissue Eng. Part A 2010, 16, 2331–2342. [Google Scholar] [CrossRef]
- Jeon, R.H.; Lee, W.J.; Son, Y.B.; Bharti, D.; Shivakumar, S.B.; Lee, S.L.; Rho, G.J. PPIA, HPRT1, and YWHAZ Genes Are Suitable for Normalization of mRNA Expression in Long-Term Expanded Human Mesenchymal Stem Cells. Biomed. Res. Int. 2019, 2019, 3093545. [Google Scholar] [CrossRef]
- Ayanoğlu, F.B.; Elçin, A.E.; Elçin, Y.M. Evaluation of the stability of standard reference genes of adipose-derived mesenchymal stem cells during in vitro proliferation and differentiation. Mol. Biol. Rep. 2020, 47, 2109–2122. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.; Hellemans, J.; Huggett, J.F.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [Green Version]
- Kowal, J.M.; Schmal, H.; Halekoh, U.; Hjelmborg, J.B.; Kassem, M. Single-cell high content imaging parameters predict functional phenotype of cultured human bone marrow stromal stem cells. Stem Cells Transl. Med. 2019, 9, 189–202. [Google Scholar] [CrossRef] [Green Version]
- Robb, K.P.; Fitzgerald, J.C.; Barry, F.; Viswanathan, S. Mesenchymal stromal cell therapy: Progress in manufacturing and assessments of potency. Cytotherapy 2019, 21, 289–306. [Google Scholar] [CrossRef] [PubMed]
- Christy, B.A.; Herzig, M.C.; Delavan, C.P.; Abaasah, I.; Cantu, C.; Salgado, C.; Lovelace, S.; Garcia, L.; Jensen, K.; Montgomery, R.; et al. Use of multiple potency assays to evaluate human mesenchymal stromal cells. J. Trauma Acute Care Surg. 2020, 89, S109–S117. [Google Scholar] [CrossRef] [PubMed]
- Rebelatto, C.K.; Aguiar, A.M.; Moretão, M.P.; Senegaglia, A.C.; Hansen, P.; Barchiki, F.; Oliveira, J.M.; Martins, J.; Kuligovski, C.; Mansur, F.; et al. Dissimilar differentiation of mesenchymal stem cells from bone marrow, umbilical cord blood, and adipose tissue. Exp. Biol. Med. 2008, 233, 901–913. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.-H.K.; Ogando, C.R.; Wang See, C.; Chang, T.-Y.; Barabino, G.A. Changes in phenotype and differentiation potential of human mesenchymal stem cells aging in vitro. Stem Cell Res. Ther. 2018, 9, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Christodoulou, I.; Kolisis, F.N.; Papaevangeliou, D.; Zoumpourlis, V. Comparative evaluation of human mesenchymal stem cells of fetal (Wharton’s jelly) and adult (adipose tissue) origin during prolonged in vitro expansion: Considerations for cytotherapy. Stem Cells Int. 2013, 2013, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Prins, H.J.; Rozemuller, H.; Vonk-Griffioen, S.; Verweij, V.G.; Dhert, W.J.; Slaper-Cortenbach, I.C.; Martens, A.C. Bone-forming capacity of mesenchymal stromal cells when cultured in the presence of human platelet lysate as substitute for fetal bovine serum. Tissue Eng. Part A 2009, 15, 3741–3751. [Google Scholar] [CrossRef]
- Ahmed, E.; Ziada, H.; Soliman, M.; El-Mezayen, H.; Hamed, S. Comparing the osteogenic potency of mesenchymal stem cells derived from bone marrow and adipose tissue. Stem Cell Adv. Res. Ther. 2020. [Google Scholar] [CrossRef]
- Janicki, P.; Boeuf, S.; Steck, E.; Egermann, M.; Kasten, P.; Richter, W. Prediction of in vivo bone forming potency of bone marrow-derived human mesenchymal stem cells. Eur. Cell Mater. 2011, 21, 488–507. [Google Scholar] [CrossRef]
- Sproul, E.P.; Argraves, W.S. A cytokine axis regulates elastin formation and degradation. Matrix Biol. 2013, 32, 86–94. [Google Scholar] [CrossRef]
- Sudo, R.; Sato, F.; Azechi, T.; Wachi, H. MiR-29-mediated elastin down-regulation contributes to inorganic phosphorus-induced osteoblastic differentiation in vascular smooth muscle cells. Genes Cells 2015, 20, 1077–1087. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.J.; Kang, S.W.; Do, H.J.; Han, I.; Shin, D.A.; Kim, J.H.; Lee, S.H. Enhancement of bone regeneration by gene delivery of BMP2/Runx2 bicistronic vector into adipose-derived stromal cells. Biomaterials 2010, 31, 5652–5659. [Google Scholar] [CrossRef] [PubMed]
- Shui, C.; Spelsberg, T.C.; Riggs, B.L.; Khosla, S. Changes in Runx2/Cbfa1 expression and activity during osteoblastic differentiation of human bone marrow stromal cells. J. Bone Miner. Res. 2003, 18, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Sanford, L.P.; Ormsby, I.; Gittenberger-de Groot, A.C.; Sariola, H.; Friedman, R.; Boivin, G.P.; Cardell, E.L.; Doetschman, T. TGFbeta2 knockout mice have multiple developmental defects that are non-overlapping with other TGFbeta knockout phenotypes. Development 1997, 124, 2659–2670. [Google Scholar] [PubMed]
- Hering, S.; Isken, E.; Knabbe, C.; Janott, J.; Jost, C.; Pommer, A.; Muhr, G.; Schatz, H.; Pfeiffer, A.F. TGFbeta1 and TGFbeta2 mRNA and protein expression in human bone samples. Exp. Clin. Endocrinol. Diabetes 2001, 109, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Kazimierczak, P.; Syta, E.; Przekora, A.; Ginalska, G. Comparison of osteogenic differentiation ability between bone marrow-derived mesenchymal stem cells and adipose tissue-derived mesenchymal stem cells. Med. Ogólna Nauk. Zdrowiu 2018, 24, 101–106. [Google Scholar] [CrossRef]
- Noël, D.; Caton, D.; Roche, S.; Bony, C.; Lehmann, S.; Casteilla, L.; Jorgensen, C.; Cousin, B. Cell specific differences between human adipose-derived and mesenchymal-stromal cells despite similar differentiation potentials. Exp. Cell Res. 2008, 314, 1575–1584. [Google Scholar] [CrossRef]
- Kulterer, B.; Friedl, G.; Jandrositz, A.; Sanchez-Cabo, F.; Prokesch, A.; Paar, C.; Scheideler, M.; Windhager, R.; Preisegger, K.H.; Trajanoski, Z. Gene expression profiling of human mesenchymal stem cells derived from bone marrow during expansion and osteoblast differentiation. BMC Genom. 2007, 8, 70. [Google Scholar] [CrossRef] [Green Version]
- Gubbiotti, M.A.; Vallet, S.D.; Ricard-Blum, S.; Iozzo, R.V. Decorin interacting network: A comprehensive analysis of decorin-binding partners and their versatile functions. Matrix Biol. 2016, 55, 7–21. [Google Scholar] [CrossRef]
- Hildebrand, A.; Romarís, M.; Rasmussen, L.M.; Heinegård, D.; Twardzik, D.R.; Border, W.A.; Ruoslahti, E. Interaction of the small interstitial proteoglycans biglycan, decorin and fibromodulin with transforming growth factor beta. Biochem. J. 1994, 302, 527–534. [Google Scholar] [CrossRef]
- Markmann, A.; Hausser, H.; Schönherr, E.; Kresse, H. Influence of decorin expression on transforming growth factor-beta-mediated collagen gel retraction and biglycan induction. Matrix Biol. 2000, 19, 631–636. [Google Scholar] [CrossRef]
- Ferdous, Z.; Wei, V.M.; Iozzo, R.; Höök, M.; Grande-Allen, K.J. Decorin-transforming growth factor- interaction regulates matrix organization and mechanical characteristics of three-dimensional collagen matrices. J. Biol. Chem. 2007, 282, 35887–35898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goetsch, K.P.; Niesler, C.U. The extracellular matrix regulates the effect of decorin and transforming growth factor beta-2 (TGF-β2) on myoblast migration. Biochem. Biophys. Res. Commun. 2016, 479, 351–357. [Google Scholar] [CrossRef] [PubMed]
- Schönherr, E.; Broszat, M.; Brandan, E.; Bruckner, P.; Kresse, H. Decorin core protein fragment Leu155-Val260 interacts with TGF-β but does not compete for decorin binding to type I collagen. Arch. Biochem. Biophys. 1998, 355, 241–248. [Google Scholar] [CrossRef]
- Erlebacher, A.; Derynck, R. Increased expression of TGF-beta 2 in osteoblasts results in an osteoporosis-like phenotype. J. Cell Biol. 1996, 132, 195–210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banfi, A.; Muraglia, A.; Dozin, B.; Mastrogiacomo, M.; Cancedda, R.; Quarto, R. Proliferation kinetics and differentiation potential of ex vivo expanded human bone marrow stromal cells: Implications for their use in cell therapy. Exp. Hematol. 2000, 28, 707–715. [Google Scholar] [CrossRef]
- Liu, J.; Ding, Y.; Liu, Z.; Liang, X. Senescence in Mesenchymal Stem Cells: Functional Alterations, Molecular Mechanisms, and Rejuvenation Strategies. Front. Cell Dev. Biol. 2020, 8, 258. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, H.; Nakatsuka, R.; Matsuoka, Y.; Sumide, K.; Fujioka, T.; Asano, H.; Iida, H.; Sonoda, Y. TGF-β Signaling Accelerates Senescence of Human Bone-Derived CD271 and SSEA-4 Double-Positive Mesenchymal Stromal Cells. Stem Cell Rep. 2018, 10, 920–932. [Google Scholar] [CrossRef] [Green Version]
- Walenda, G.; Abnaof, K.; Joussen, S.; Meurer, S.; Smeets, H.; Rath, B.; Hoffmann, K.; Fröhlich, H.; Zenke, M.; Weiskirchen, R.; et al. TGF-beta1 does not induce senescence of multipotent mesenchymal stromal cells and has similar effects in early and late passages. PLoS ONE 2013, 8, e77656. [Google Scholar] [CrossRef] [Green Version]
- Gibaja, A.; Aburto, M.R.; Pulido, S.; Collado, M.; Hurle, J.M.; Varela-Nieto, I.; Magariños, M. TGFβ2-induced senescence during early inner ear development. Sci. Rep. 2019, 9, 5912. [Google Scholar] [CrossRef] [Green Version]
- Schellenberg, A.; Mauen, S.; Koch, C.M.; Jans, R.; de Waele, P.; Wagner, W. Proof of principle: Quality control of therapeutic cell preparations using senescence-associated DNA-methylation changes. BMC Res. Notes 2014, 7, 254. [Google Scholar] [CrossRef] [Green Version]
- de Almeida, D.C.; Ferreira, M.R.; Franzen, J.; Weidner, C.I.; Frobel, J.; Zenke, M.; Costa, I.G.; Wagner, W. Epigenetic Classification of Human Mesenchymal Stromal Cells. Stem Cell Rep. 2016, 6, 168–175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mankani, M.H.; Kuznetsov, S.A.; Marshall, G.W.; Robey, P.G. Creation of new bone by the percutaneous injection of human bone marrow stromal cell and HA/TCP suspensions. Tissue Eng. Part A 2008, 14, 1949–1958. [Google Scholar] [CrossRef] [PubMed]
- Haffner-Luntzer, M.; Kovtun, A.; Rapp, A.E.; Ignatius, A. Mouse Models in Bone Fracture Healing Research. Curr. Mol. Biol. Rep. 2016, 2, 101–111. [Google Scholar] [CrossRef] [Green Version]
- Samsonraj, R.M.; Rai, B.; Sathiyanathan, P.; Puan, K.J.; Rötzschke, O.; Hui, J.H.; Raghunath, M.; Stanton, L.W.; Nurcombe, V.; Cool, S.M. Establishing criteria for human mesenchymal stem cell potency. Stem Cells 2015, 33, 1878–1891. [Google Scholar] [CrossRef]
- Xiao, B.; Wang, G.; Li, W. Weighted gene correlation network analysis reveals novel biomarkers associated with mesenchymal stromal cell differentiation in early phase. PeerJ 2020, 8, e8907. [Google Scholar] [CrossRef] [Green Version]
- Sathiyanathan, P.; Samsonraj, R.M.; Tan, C.L.L.; Ling, L.; Lezhava, A.; Nurcombe, V.; Stanton, L.W.; Cool, S.M. A genomic biomarker that identifies human bone marrow-derived mesenchymal stem cells with high scalability. Stem Cells 2020. [Google Scholar] [CrossRef]
- Tsai, A.-C.; Jeske, R.; Chen, X.; Yuan, X.; Li, Y. Influence of Microenvironment on Mesenchymal Stem Cell Therapeutic Potency: From Planar Culture to Microcarriers. Front. Bioeng. Biotechnol. 2020, 8, 640. [Google Scholar] [CrossRef]










| Gene Name | Primer Sequence | Amplified Length (bp) |
|---|---|---|
| Alkaline phosphatase (ALPL) | 5’-GATGTGGAGTATGAGAGTGACG-3’ (sense) 5’-GGTCAAGGGTCAGGAGTTC-3’ (antisense) | 142 |
| Collagen type I, alpha 1 (COL1A2) | 5’-AGGACAAGAAACACGTCTGG-3’ (sense) 5’-GGTGATGTTCTGAGAGGCATAG-3’ (antisense) | 146 |
| Decorin (DCN) | 5’-AAAATGCCCAAAACTCTTCAGG-3’ (sense) 5’-GCCCCATTTTCAATTCCTGAG-3’ (antisense) | 146 |
| Elastin (ELN) | 5’-CCTGGCTTCGGATTGTCTC-3’ (sense) 5’-CAAAGGGTTTACATTCTCCACC-3’ (antisense) | 148 |
| Runt related transcription factor 2 (RUNX2) | 5’-TTCACCTTGACCATAACCGTC-3’ (sense) 5’-GGCGGTCAGAGAACAAACTAG-3’ (antisense) | 148 |
| Transforming growth factor beta 2 (TGFB2) | 5’-CAAAATAGACATGCCGCCCTTC-3’ (sense) 5’-GAAGGGCGGCATGTCTATTTTG-3’ (antisense) | 150 |
| Actin beta (ACTB) | 5’-ACCTTCTACAATGAGCTGCG-3’ (sense) 5’-CCTGGATAGCAACGTACATGG-3’ (antisense) | 148 |
| Glucuronidase beta (GUSB) | 5’-CCAAGGGTTACTTTGTCCAGA-3’ (sense) 5’-TAATTCACCAGCCCACTGTC-3’ (antisense) | 151 |
| Tyrosine 3 monooxygenase/tryptophan 5-monooxygenase activation protein zeta (YWHAZ) | 5’-TGACAAGAAAGGGATTGTCGAT-3’ (sense) 5’-TCTGGGGAGTTCAGAATCTCAT-3’ (antisense) | 150 |
| Gene Name | Protein Function/Gene Group | TaqMan Assay |
|---|---|---|
| Alkaline phosphatase (ALPL) | Phosphatase (bone marker, gene of interest) | Hs01029144_m1 |
| Collagen type I, alpha 1 (COL1A2) | Extracellular matrix structural protein (bone marker, gene of interest) | Hs01028956_m1 |
| Decorin (DCN) | Collagen associated proteoglycan (bone marker, gene of interest) | Hs00370385_m1 |
| Elastin (ELN) | Extracellular matrix structural protein (bone marker, gene of interest) | Hs00355783_m1 |
| Runt related transcription factor 2 (RUNX2) | Transcription factor (bone marker, gene of interest) | Hs01047973_m1 |
| Actin beta (ACTB) | Structural protein (housekeeping gene/control) | Hs99999903_m1 |
| Glucuronidase beta (GUSB) | Lysosomal enzyme (housekeeping gene/control) | Hs00939627_m1 |
| Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta (YWHAZ) | Signalling pathway protein (housekeeping gene/control) | Hs00237047_m1 |
| Donor | MSC Source | Passage of Cryopreserved hMSC (n) | Passage Number for In Vitro mRNA Harvest | Passage Number before In Vivo Heterotopic Implantation | Previously Quantified In Vivo Heterotopic Osteoid/Bone (%) ([18]) | Bone Formation In Vivo (Present Study) |
|---|---|---|---|---|---|---|
| #1 | BM | 5 | 8 | 9 | 15.3 | ND † |
| #2 | BM | 4 | 7 | 8 | 18.8 | yes |
| #3 | BM | 4 | 7 | 8 | 18.1 | yes |
| #4 | BM | 3 | 6 | 7 | 0.1 | no |
| #5 | BM | 1 | 5 | 6 | 5.4 | no |
| #6 | BM | 3 | 6 | 7 | 15.3 | yes |
| #7 | AT | 1 | 4 | 5 | ND | yes |
| #8 | AT | 5 | 8 | 9 | ND | yes |
| #9 | AT | 4 | 7 | 8 | ND | yes |
| SYBR | MAD 1W | A/G/Y | A/G | A/Y | G/Y | A | G | Y |
|---|---|---|---|---|---|---|---|---|
| ALPL | 4.546 | 4.751 | 6.007 | 6.007 | 7.184 | 1.508 | 3.412 | |
| COL1A2 | 6.944 | 6.854 | 6.778 | 6.973 | 6.703 | 6.997 | 7.127 | |
| DCN | 13.825 | 9.343 | 14.523 | 3.284 | 31.250 | 2.792 | 12.863 | |
| ELN | 7.391 | 7.392 | 7.390 | 7.389 | 5.746 | 5.749 | 6.934 | |
| RUNX2 | 5.198 | 5.661 | 5.146 | 5.406 | 9.837 | 1.759 | 4.441 | |
| TGFΒ2 | 6.364 | 6.508 | 6.438 | 6.299 | 3.754 | 5.125 | 5.087 | |
| AVERAGE MAD | 7.378 | 6.752 | 7.714 | 5.893 | 10.746 | 3.988 | 6.644 | |
| MAD 2W | A/G/Y | A/G | A/Y | G/Y | A | G | Y | |
| ALPL | 5.264 | 8.523 | 3.351 | 4.112 | 5.388 | 0.828 | 4.661 | |
| COL1A2 | 11.575 | 11.623 | 11.464 | 11.641 | 11.167 | 11.383 | 11.640 | |
| DCN | 21.285 | 9.018 | 22.687 | 5.132 | 29.062 | 4.576 | 15.944 | |
| ELN | 11.908 | 11.908 | 11.906 | 11.909 | 9.490 | 9.495 | 11.452 | |
| RUNX2 | 10.717 | 11.065 | 10.069 | 11.020 | 7.547 | 7.697 | 10.116 | |
| TGFΒ2 | 8.966 | 9.050 | 10.541 | 9.633 | 8.134 | 7.261 | 10.001 | |
| AVERAGE MAD | 11.619 | 10.198 | 11.670 | 8.908 | 11.798 | 6.874 | 10.636 | |
| TaqMan | MAD 1W | A/G/Y | A/G | A/Y | G/Y | A | G | Y |
| ALPL | 2.879 | 3.553 | 3.553 | 3.553 | 5.135 | 4.113 | 3.899 | |
| COL1A2 | 5.530 | 5.546 | 5.546 | 5.598 | 5.301 | 5.684 | 5.490 | |
| DCN | 13.710 | 11.984 | 11.984 | 12.322 | 20.311 | 6.782 | 19.807 | |
| ELN | 5.961 | 5.962 | 5.962 | 5.962 | 5.963 | 5.964 | 5.960 | |
| RUNX2 | 4.564 | 4.616 | 4.616 | 5.054 | 4.276 | 5.355 | 4.765 | |
| AVERAGE MAD | 6.529 | 6.332 | 6.332 | 6.498 | 8.197 | 5.580 | 7.984 | |
| MAD 2W | A/G/Y | A/G | A/Y | G/Y | A | G | Y | |
| ALPL | 3.479 | 2.817 | 3.192 | 4.249 | 5.986 | 4.574 | 4.527 | |
| COL1A2 | 6.364 | 6.374 | 6.247 | 6.409 | 6.006 | 6.515 | 6.305 | |
| DCN | 9.446 | 8.636 | 14.352 | 7.316 | 21.372 | 4.217 | 10.651 | |
| ELN | 6.703 | 6.704 | 6.702 | 6.704 | 6.705 | 6.706 | 6.704 | |
| RUNX2 | 5.670 | 5.267 | 5.243 | 5.813 | 4.777 | 5.988 | 5.379 | |
| AVERAGE MAD | 6.332 | 5.959 | 7.147 | 6.098 | 8.969 | 5.600 | 6.713 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ofiteru, A.M.; Becheru, D.F.; Gharbia, S.; Balta, C.; Herman, H.; Mladin, B.; Ionita, M.; Hermenean, A.; Burns, J.S. Qualifying Osteogenic Potency Assay Metrics for Human Multipotent Stromal Cells: TGF-β2 a Telling Eligible Biomarker. Cells 2020, 9, 2559. https://doi.org/10.3390/cells9122559
Ofiteru AM, Becheru DF, Gharbia S, Balta C, Herman H, Mladin B, Ionita M, Hermenean A, Burns JS. Qualifying Osteogenic Potency Assay Metrics for Human Multipotent Stromal Cells: TGF-β2 a Telling Eligible Biomarker. Cells. 2020; 9(12):2559. https://doi.org/10.3390/cells9122559
Chicago/Turabian StyleOfiteru, Augustin M., Diana F. Becheru, Sami Gharbia, Cornel Balta, Hildegard Herman, Bianca Mladin, Mariana Ionita, Anca Hermenean, and Jorge S. Burns. 2020. "Qualifying Osteogenic Potency Assay Metrics for Human Multipotent Stromal Cells: TGF-β2 a Telling Eligible Biomarker" Cells 9, no. 12: 2559. https://doi.org/10.3390/cells9122559