MicroRNA-Based Discovery of Biomarkers, Therapeutic Targets, and Repositioning Drugs for Breast Cancer
Abstract
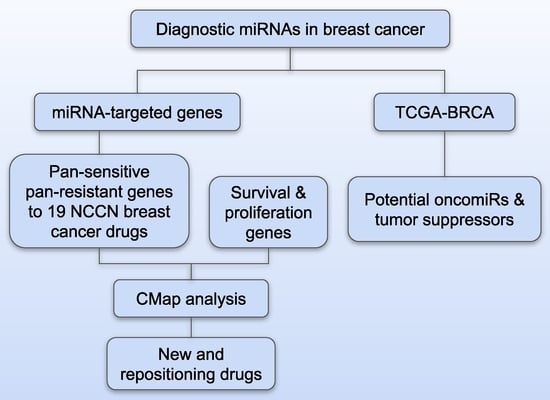
:1. Introduction
2. Materials and Methods
2.1. Patient Samples
2.2. RNA Isolation and Quality Assessment
2.3. Microarray Analysis
2.4. The Cancer Genome Atlas (TCGA) Breast Cancer Patient Cohort
2.5. Proliferation Assays
2.6. Cancer Cell Line Encyclopedia (CCLE)
2.7. Drug Sensitivity Data of Breast Cancer Cell Lines
2.8. Bioinformatic Tools
2.9. Statistical Analysis
3. Results
3.1. Differential MiRNA Expression in Breast Cancer Tissue and Blood
3.2. Association with Drug Sensitivity
3.3. Discovery of Potential Therapeutic Targets and Repositioning Drugs
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Breast Cancer-Metastatic: Statistics. Available online: https://www.cancer.net/cancer-types/breast-cancer-metastatic/statistics (accessed on 9 June 2023).
- Paik, S.; Shak, S.; Tang, G.; Kim, C.; Baker, J.; Cronin, M.; Baehner, F.L.; Walker, M.G.; Watson, D.; Park, T.; et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N. Engl. J. Med. 2004, 351, 2817–2826. [Google Scholar]
- van de Vijver, M.J.; He, Y.D.; van‘t Veer, L.J.; Dai, H.; Hart, A.A.; Voskuil, D.W.; Schreiber, G.J.; Peterse, J.L.; Roberts, C.; Marton, M.J.; et al. A gene-expression signature as a predictor of survival in breast cancer. N. Engl. J. Med. 2002, 347, 1999–2009. [Google Scholar]
- van’t Veer, L.J.; Dai, H.; van de Vijver, M.J.; He, Y.D.; Hart, A.A.; Mao, M.; Peterse, H.L.; van der, K.K.; Marton, M.J.; Witteveen, A.T.; et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature 2002, 415, 530–536. [Google Scholar]
- Parker, J.S.; Mullins, M.; Cheang, M.C.; Leung, S.; Voduc, D.; Vickery, T.; Davies, S.; Fauron, C.; He, X.; Hu, Z.; et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2009, 27, 1160–1167. [Google Scholar] [CrossRef]
- Prat, A.; Pineda, E.; Adamo, B.; Galván, P.; Fernández, A.; Gaba, L.; Díez, M.; Viladot, M.; Arance, A.; Muñoz, M. Clinical implications of the intrinsic molecular subtypes of breast cancer. Breast 2015, 24 (Suppl. S2), S26–S35. [Google Scholar] [CrossRef] [Green Version]
- Mariotto, A.B.; Etzioni, R.; Hurlbert, M.; Penberthy, L.; Mayer, M. Estimation of the Number of Women Living with Metastatic Breast Cancer in the United States. Cancer Epidemiol. Biomark. Prev. 2017, 26, 809–815. [Google Scholar] [CrossRef] [Green Version]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [Green Version]
- Kucherenko, M.M.; Shcherbata, H.R. miRNA targeting and alternative splicing in the stress response–events hosted by membrane-less compartments. J. Cell Sci. 2018, 131, jcs202002. [Google Scholar]
- Wang, K.; Zhang, S.; Weber, J.; Baxter, D.; Galas, D.J. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 2010, 38, 7248–7259. [Google Scholar] [PubMed] [Green Version]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [Green Version]
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Yu, F.; Ding, H.; Wang, Y.; Li, P.; Wang, K. Emerging Function and Clinical Values of Exosomal MicroRNAs in Cancer. Mol. Ther. Nucleic Acids 2019, 16, 791–804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farazi, T.A.; Spitzer, J.I.; Morozov, P.; Tuschl, T. miRNAs in human cancer. J. Pathol. 2011, 223, 102–115. [Google Scholar] [CrossRef] [PubMed]
- Ferracin, M.; Veronese, A.; Negrini, M. Micromarkers: miRNAs in cancer diagnosis and prognosis. Expert Rev. Mol. Diagn. 2010, 10, 297–308. [Google Scholar] [CrossRef]
- Iorio, M.V.; Croce, C.M. microRNA involvement in human cancer. Carcinogenesis 2012, 33, 1126–1133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blenkiron, C.; Goldstein, L.D.; Thorne, N.P.; Spiteri, I.; Chin, S.-F.; Dunning, M.J.; Barbosa-Morais, N.L.; Teschendorff, A.E.; Green, A.R.; Ellis, I.O. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 2007, 8, R214. [Google Scholar] [CrossRef] [Green Version]
- Iorio, M.V.; Ferracin, M.; Liu, C.-G.; Veronese, A.; Spizzo, R.; Sabbioni, S.; Magri, E.; Pedriali, M.; Fabbri, M.; Campiglio, M. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005, 65, 7065–7070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nelson, P.T.; Baldwin, D.A.; Scearce, L.M.; Oberholtzer, J.C.; Tobias, J.W.; Mourelatos, Z. Microarray-based, high-throughput gene expression profiling of microRNAs. Nat. Methods 2004, 1, 155–161. [Google Scholar] [CrossRef]
- Shingara, J.; Keiger, K.; Shelton, J.; Laosinchai-Wolf, W.; Powers, P.; Conrad, R.; Brown, D.; Labourier, E. An optimized isolation and labeling platform for accurate microRNA expression profiling. Rna 2005, 11, 1461–1470. [Google Scholar] [CrossRef] [Green Version]
- Cortez, M.A.; Bueso-Ramos, C.; Ferdin, J.; Lopez-Berestein, G.; Sood, A.K.; Calin, G.A. MicroRNAs in body fluids—The mix of hormones and biomarkers. Nat. Rev. Clin. Oncol. 2011, 8, 467–477. [Google Scholar] [CrossRef] [Green Version]
- Zhao, H.; Shen, J.; Medico, L.; Wang, D.; Ambrosone, C.B.; Liu, S. A pilot study of circulating miRNAs as potential biomarkers of early stage breast cancer. PLoS ONE 2010, 5, e13735. [Google Scholar]
- Baldasici, O.; Pileczki, V.; Cruceriu, D.; Gavrilas, L.I.; Tudoran, O.; Balacescu, L.; Vlase, L.; Balacescu, O. Breast Cancer-Delivered Exosomal miRNA as Liquid Biopsy Biomarkers for Metastasis Prediction: A Focus on Translational Research with Clinical Applicability. Int. J. Mol. Sci. 2022, 23, 9371. [Google Scholar] [CrossRef] [PubMed]
- Huang, W. MicroRNAs: Biomarkers, diagnostics, and therapeutics. Bioinform. MicroRNA Res. 2017, 57–67. [Google Scholar]
- Diener, C.; Keller, A.; Meese, E. Emerging concepts of miRNA therapeutics: From cells to clinic. Trends Genet. 2022, 38, 613–626. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Narayan, R.; Corsello, S.M.; Peck, D.D.; Natoli, T.E.; Lu, X.; Gould, J.; Davis, J.F.; Tubelli, A.A.; Asiedu, J.K.; et al. A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell 2017, 171, 1437–1452.e1417. [Google Scholar] [CrossRef]
- Lamb, J.; Crawford, E.D.; Peck, D.; Modell, J.W.; Blat, I.C.; Wrobel, M.J.; Lerner, J.; Brunet, J.P.; Subramanian, A.; Ross, K.N.; et al. The Connectivity Map: Using gene-expression signatures to connect small molecules, genes, and disease. Science 2006, 313, 1929–1935. [Google Scholar] [CrossRef] [Green Version]
- Beekman, J.M.; Reischl, J.; Henderson, D.; Bauer, D.; Ternes, R.; Pena, C.; Lathia, C.; Heubach, J.F. Recovery of microarray-quality RNA from frozen EDTA blood samples. J. Pharmacol. Toxicol. Methods 2009, 59, 44–49. [Google Scholar] [CrossRef]
- Vasaikar, S.V.; Straub, P.; Wang, J.; Zhang, B. LinkedOmics: Analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018, 46, D956–D963. [Google Scholar] [CrossRef] [Green Version]
- Dempster, J.M.; Rossen, J.; Kazachkova, M.; Pan, J.; Kugener, G.; Root, D.E.; Tsherniak, A. Extracting Biological Insights from the Project Achilles Genome-Scale CRISPR Screens in Cancer Cell Lines. bioRxiv 2019. [Google Scholar] [CrossRef]
- Meyers, R.M.; Bryan, J.G.; McFarland, J.M.; Weir, B.A.; Sizemore, A.E.; Xu, H.; Dharia, N.V.; Montgomery, P.G.; Cowley, G.S.; Pantel, S.; et al. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat. Genet. 2017, 49, 1779–1784. [Google Scholar] [CrossRef] [Green Version]
- Behan, F.M.; Iorio, F.; Picco, G.; Goncalves, E.; Beaver, C.M.; Migliardi, G.; Santos, R.; Rao, Y.; Sassi, F.; Pinnelli, M.; et al. Prioritization of cancer therapeutic targets using CRISPR-Cas9 screens. Nature 2019, 568, 511–516. [Google Scholar] [CrossRef]
- Nusinow, D.P.; Szpyt, J.; Ghandi, M.; Rose, C.M.; McDonald, E.R., 3rd; Kalocsay, M.; Jané-Valbuena, J.; Gelfand, E.; Schweppe, D.K.; Jedrychowski, M.; et al. Quantitative Proteomics of the Cancer Cell Line Encyclopedia. Cell 2020, 180, 387–402.e316. [Google Scholar] [CrossRef] [PubMed]
- Corsello, S.M.; Nagari, R.T.; Spangler, R.D.; Rossen, J.; Kocak, M.; Bryan, J.G.; Humeidi, R.; Peck, D.; Wu, X.; Tang, A.A.; et al. Discovering the anti-cancer potential of non-oncology drugs by systematic viability profiling. Nat. Cancer 2020, 1, 235–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, W.; Soares, J.; Greninger, P.; Edelman, E.J.; Lightfoot, H.; Forbes, S.; Bindal, N.; Beare, D.; Smith, J.A.; Thompson, I.R.; et al. Genomics of Drug Sensitivity in Cancer (GDSC): A resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013, 41, D955–D961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iorio, F.; Knijnenburg, T.A.; Vis, D.J.; Bignell, G.R.; Menden, M.P.; Schubert, M.; Aben, N.; Goncalves, E.; Barthorpe, S.; Lightfoot, H.; et al. A Landscape of Pharmacogenomic Interactions in Cancer. Cell 2016, 166, 740–754. [Google Scholar] [CrossRef] [Green Version]
- Garnett, M.J.; Edelman, E.J.; Heidorn, S.J.; Greenman, C.D.; Dastur, A.; Lau, K.W.; Greninger, P.; Thompson, I.R.; Luo, X.; Soares, J.; et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature 2012, 483, 570–575. [Google Scholar] [CrossRef] [Green Version]
- Ye, Q.; Mohamed, R.; Dukhlallah, D.; Gencheva, M.; Hu, G.; Pearce, M.C.; Kolluri, S.K.; Marsh, C.B.; Eubank, T.D.; Ivanov, A.V.; et al. Molecular Analysis of ZNF71 KRAB in Non-Small-Cell Lung Cancer. Int. J. Mol. Sci. 2021, 22, 3752. [Google Scholar] [CrossRef]
- Ye, Q.; Singh, S.; Qian, P.R.; Guo, N.L. Immune-Omics Networks of CD27, PD1, and PDL1 in Non-Small Cell Lung Cancer. Cancers 2021, 13, 4296. [Google Scholar] [CrossRef]
- Vlachos, I.S.; Paraskevopoulou, M.D.; Karagkouni, D.; Georgakilas, G.; Vergoulis, T.; Kanellos, I.; Anastasopoulos, I.L.; Maniou, S.; Karathanou, K.; Kalfakakou, D.; et al. DIANA-TarBase v7.0: Indexing more than half a million experimentally supported miRNA:mRNA interactions. Nucleic Acids Res. 2015, 43, D153–D159. [Google Scholar] [CrossRef]
- Karagkouni, D.; Paraskevopoulou, M.D.; Chatzopoulos, S.; Vlachos, I.S.; Tastsoglou, S.; Kanellos, I.; Papadimitriou, D.; Kavakiotis, I.; Maniou, S.; Skoufos, G.; et al. DIANA-TarBase v8: A decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Res. 2018, 46, D239–D245. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Bardes, E.E.; Aronow, B.J.; Jegga, A.G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009, 37, W305–W311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome research 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Volinia, S.; Galasso, M.; Sana, M.E.; Wise, T.F.; Palatini, J.; Huebner, K.; Croce, C.M. Breast cancer signatures for invasiveness and prognosis defined by deep sequencing of microRNA. Proc. Natl. Acad. Sci. USA 2012, 109, 3024–3029. [Google Scholar] [CrossRef] [PubMed]
- Petrelli, A.; Bellomo, S.E.; Sarotto, I.; Kubatzki, F.; Sgandurra, P.; Maggiorotto, F.; Di Virgilio, M.R.; Ponzone, R.; Geuna, E.; Galizia, D.; et al. MiR-100 is a predictor of endocrine responsiveness and prognosis in patients with operable luminal breast cancer. ESMO Open 2020, 5, e000937. [Google Scholar] [CrossRef]
- Xie, H.; Xiao, R.; He, Y.; He, L.; Xie, C.; Chen, J.; Hong, Y. MicroRNA-100 inhibits breast cancer cell proliferation, invasion and migration by targeting FOXA1. Oncol. Lett. 2021, 22, 816. [Google Scholar] [CrossRef]
- Normann, L.S.; Haugen, M.H.; Aure, M.R.; Kristensen, V.N.; Mælandsmo, G.M.; Sahlberg, K.K. miR-101-5p Acts as a Tumor Suppressor in HER2-Positive Breast Cancer Cells and Improves Targeted Therapy. Breast Cancer Dove Med. Press 2022, 14, 25–39. [Google Scholar] [CrossRef]
- Wang, L.; Li, L.; Guo, R.; Li, X.; Lu, Y.; Guan, X.; Gitau, S.C.; Wang, L.; Xu, C.; Yang, B.; et al. miR-101 promotes breast cancer cell apoptosis by targeting Janus kinase 2. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2014, 34, 413–422. [Google Scholar] [CrossRef]
- Hong, B.S.; Ryu, H.S.; Kim, N.; Kim, J.; Lee, E.; Moon, H.; Kim, K.H.; Jin, M.S.; Kwon, N.H.; Kim, S.; et al. Tumor Suppressor miRNA-204-5p Regulates Growth, Metastasis, and Immune Microenvironment Remodeling in Breast Cancer. Cancer Res. 2019, 79, 1520–1534. [Google Scholar] [CrossRef]
- Liang, W.H.; Li, N.; Yuan, Z.Q.; Qian, X.L.; Wang, Z.H. DSCAM-AS1 promotes tumor growth of breast cancer by reducing miR-204-5p and up-regulating RRM2. Mol. Carcinog. 2019, 58, 461–473. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, L.; Xu, X.; He, X.; Wang, G.; Fan, C.; Zheng, Q.; Li, F. miR-205/RunX2 axis negatively regulates CD44(+)/CD24(-) breast cancer stem cell activity. Am. J. Cancer Res. 2020, 10, 1871–1887. [Google Scholar]
- Plantamura, I.; Cataldo, A.; Cosentino, G.; Iorio, M.V. miR-205 in Breast Cancer: State of the Art. Int. J. Mol. Sci. 2020, 22, 27. [Google Scholar] [CrossRef] [PubMed]
- Croset, M.; Pantano, F.; Kan, C.W.S.; Bonnelye, E.; Descotes, F.; Alix-Panabières, C.; Lecellier, C.H.; Bachelier, R.; Allioli, N.; Hong, S.S.; et al. miRNA-30 Family Members Inhibit Breast Cancer Invasion, Osteomimicry, and Bone Destruction by Directly Targeting Multiple Bone Metastasis-Associated Genes. Cancer Res. 2018, 78, 5259–5273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Brien, K.P.; Khan, S.; Gilligan, K.E.; Zafar, H.; Lalor, P.; Glynn, C.; O’Flatharta, C.; Ingoldsby, H.; Dockery, P.; De Bhulbh, A.; et al. Employing mesenchymal stem cells to support tumor-targeted delivery of extracellular vesicle (EV)-encapsulated microRNA-379. Oncogene 2018, 37, 2137–2149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, S.; Brougham, C.L.; Ryan, J.; Sahrudin, A.; O’Neill, G.; Wall, D.; Curran, C.; Newell, J.; Kerin, M.J.; Dwyer, R.M. miR-379 regulates cyclin B1 expression and is decreased in breast cancer. PLoS ONE 2013, 8, e68753. [Google Scholar] [CrossRef] [Green Version]
- Pollari, S.; Leivonen, S.K.; Perälä, M.; Fey, V.; Käkönen, S.M.; Kallioniemi, O. Identification of microRNAs inhibiting TGF-β-induced IL-11 production in bone metastatic breast cancer cells. PLoS ONE 2012, 7, e37361. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Zhu, Q.; Tang, L. MiR-99a antitumor activity in human breast cancer cells through targeting of mTOR expression. PLoS ONE 2014, 9, e92099. [Google Scholar] [CrossRef] [Green Version]
- Qin, H.; Liu, W. MicroRNA-99a-5p suppresses breast cancer progression and cell-cycle pathway through downregulating CDC25A. J. Cell. Physiol. 2019, 234, 3526–3537. [Google Scholar] [CrossRef]
- Garrido-Cano, I.; Adam-Artigues, A.; Lameirinhas, A.; Blandez, J.F.; Candela-Noguera, V.; Rojo, F.; Zazo, S.; Madoz-Gúrpide, J.; Lluch, A.; Bermejo, B.; et al. miR-99a-5p modulates doxorubicin resistance via the COX-2/ABCG2 axis in triple-negative breast cancer: From the discovery to in vivo studies. Cancer Commun. 2022, 42, 1412–1416. [Google Scholar] [CrossRef]
- Tang, Y.; Yang, S.; Wang, M.; Liu, D.; Liu, Y.; Zhang, Y.; Zhang, Q. Epigenetically altered miR-193a-3p promotes HER2 positive breast cancer aggressiveness by targeting GRB7. Int. J. Mol. Med. 2019, 43, 2352–2360. [Google Scholar] [CrossRef]
- Yu, M.; Liu, Z.; Liu, Y.; Zhou, X.; Sun, F.; Liu, Y.; Li, L.; Hua, S.; Zhao, Y.; Gao, H.; et al. PTP1B markedly promotes breast cancer progression and is regulated by miR-193a-3p. FEBS J. 2019, 286, 1136–1153. [Google Scholar] [CrossRef]
- Arnesen, S.; Polaski, J.T.; Blanchard, Z.; Osborne, K.S.; Welm, A.L.; O’Connell, R.M.; Gertz, J. Estrogen receptor alpha mutations regulate gene expression and cell growth in breast cancer through microRNAs. NAR Cancer 2023, 5, zcad027. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Xi, X.; Li, J.; Ni, J.; Yang, H.; Wen, C.; Wen, M. miR-301b and NR3C2 co-regulate cells malignant properties and have the potential to be independent prognostic factors in breast cancer. J. Biochem. Mol. Toxicol. 2021, 35, e22650. [Google Scholar] [CrossRef] [PubMed]
- Lei, B.; Wang, D.; Zhang, M.; Deng, Y.; Jiang, H.; Li, Y. miR-615-3p promotes the epithelial-mesenchymal transition and metastasis of breast cancer by targeting PICK1/TGFBRI axis. J. Exp. Clin. Cancer Res. CR 2020, 39, 71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, Y.X.; Bradbury, R.; Flamini, V.; Wu, B.; Jordan, N.; Jiang, W.G. MicroRNA-7 suppresses the homing and migration potential of human endothelial cells to highly metastatic human breast cancer cells. Br. J. Cancer 2017, 117, 89–101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okuda, H.; Xing, F.; Pandey, P.R.; Sharma, S.; Watabe, M.; Pai, S.K.; Mo, Y.Y.; Iiizumi-Gairani, M.; Hirota, S.; Liu, Y.; et al. miR-7 suppresses brain metastasis of breast cancer stem-like cells by modulating KLF4. Cancer Res. 2013, 73, 1434–1444. [Google Scholar] [CrossRef] [Green Version]
- Das, S.G.; Romagnoli, M.; Mineva, N.D.; Barillé-Nion, S.; Jézéquel, P.; Campone, M.; Sonenshein, G.E. miR-720 is a downstream target of an ADAM8-induced ERK signaling cascade that promotes the migratory and invasive phenotype of triple-negative breast cancer cells. Breast Cancer Res. BCR 2016, 18, 40. [Google Scholar] [CrossRef]
- Lee, J.; Galloway, R.; Grandjean, G.; Jacob, J.; Humphries, J.; Bartholomeusz, C.; Goodstal, S.; Lim, B.; Bartholomeusz, G.; Ueno, N.T.; et al. Comprehensive Two- and Three-Dimensional RNAi Screening Identifies PI3K Inhibition as a Complement to MEK Inhibitor AS703026 for Combination Treatment of Triple-Negative Breast Cancer. J. Cancer 2015, 6, 1306–1319. [Google Scholar] [CrossRef] [Green Version]
- Vazquez-Ortiz, G.; Chisholm, C.; Xu, X.; Lahusen, T.J.; Li, C.; Sakamuru, S.; Huang, R.; Thomas, C.J.; Xia, M.; Deng, C. Drug repurposing screen identifies lestaurtinib amplifies the ability of the poly (ADP-ribose) polymerase 1 inhibitor AG14361 to kill breast cancer associated gene-1 mutant and wild type breast cancer cells. Breast Cancer Res. BCR 2014, 16, R67. [Google Scholar] [CrossRef]
- Roskoski, R., Jr. Properties of FDA-approved small molecule protein kinase inhibitors: A 2022 update. Pharmacol. Res. 2022, 175, 106037. [Google Scholar] [CrossRef]
- Kawai, M.; Nakashima, A.; Kamada, S.; Kikkawa, U. Midostaurin preferentially attenuates proliferation of triple-negative breast cancer cell lines through inhibition of Aurora kinase family. J. Biomed. Sci. 2015, 22, 48. [Google Scholar] [CrossRef] [Green Version]
- Huang, K.M.; Zavorka Thomas, M.; Magdy, T.; Eisenmann, E.D.; Uddin, M.E.; DiGiacomo, D.F.; Pan, A.; Keiser, M.; Otter, M.; Xia, S.H.; et al. Targeting OCT3 attenuates doxorubicin-induced cardiac injury. Proc. Natl. Acad. Sci. USA 2021, 118, e2020168118. [Google Scholar] [CrossRef]
- Pedersen, A.M.; Thrane, S.; Lykkesfeldt, A.E.; Yde, C.W. Sorafenib and nilotinib resensitize tamoxifen resistant breast cancer cells to tamoxifen treatment via estrogen receptor α. Int. J. Oncol. 2014, 45, 2167–2175. [Google Scholar] [CrossRef] [Green Version]
- Allen, L.F.; Sebolt-Leopold, J.; Meyer, M.B. CI-1040 (PD184352), a targeted signal transduction inhibitor of MEK (MAPKK). Semin. Oncol. 2003, 30, 105–116. [Google Scholar] [CrossRef] [PubMed]
- Rinehart, J.; Adjei, A.A.; Lorusso, P.M.; Waterhouse, D.; Hecht, J.R.; Natale, R.B.; Hamid, O.; Varterasian, M.; Asbury, P.; Kaldjian, E.P.; et al. Multicenter phase II study of the oral MEK inhibitor, CI-1040, in patients with advanced non-small-cell lung, breast, colon, and pancreatic cancer. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2004, 22, 4456–4462. [Google Scholar] [CrossRef] [PubMed]
- Mehmood, A.; Nawab, S.; Jin, Y.; Hassan, H.; Kaushik, A.C.; Wei, D.Q. Ranking Breast Cancer Drugs and Biomarkers Identification Using Machine Learning and Pharmacogenomics. ACS Pharmacol. Transl. Sci. 2023, 6, 399–409. [Google Scholar] [CrossRef] [PubMed]
- You, K.S.; Yi, Y.W.; Cho, J.; Seong, Y.S. Dual Inhibition of AKT and MEK Pathways Potentiates the Anti-Cancer Effect of Gefitinib in Triple-Negative Breast Cancer Cells. Cancers 2021, 13, 1205. [Google Scholar] [CrossRef]
- Ishay-Ronen, D.; Diepenbruck, M.; Kalathur, R.K.R.; Sugiyama, N.; Tiede, S.; Ivanek, R.; Bantug, G.; Morini, M.F.; Wang, J.; Hess, C.; et al. Gain Fat-Lose Metastasis: Converting Invasive Breast Cancer Cells into Adipocytes Inhibits Cancer Metastasis. Cancer Cell 2019, 35, 17–32.e16. [Google Scholar] [CrossRef] [Green Version]
- You, Y.; Niu, Y.; Zhang, J.; Huang, S.; Ding, P.; Sun, F.; Wang, X. U0126: Not only a MAPK kinase inhibitor. Front. Pharmacol. 2022, 13, 927083. [Google Scholar] [CrossRef] [PubMed]
- Lodi, A.; Woods, S.M.; Ronen, S.M. Treatment with the MEK inhibitor U0126 induces decreased hyperpolarized pyruvate to lactate conversion in breast, but not prostate, cancer cells. NMR Biomed. 2013, 26, 299–306. [Google Scholar] [CrossRef] [Green Version]
- Zhao, L.Y.; Huang, C.; Li, Z.F.; Liu, L.; Ni, L.; Song, T.S. STAT1/2 is involved in the inhibition of cell growth induced by U0126 in HeLa cells. Cell. Mol. Biol. 2009, 55, 1168–1174. [Google Scholar]
- Aksamitiene, E.; Kholodenko, B.N.; Kolch, W.; Hoek, J.B.; Kiyatkin, A. PI3K/Akt-sensitive MEK-independent compensatory circuit of ERK activation in ER-positive PI3K-mutant T47D breast cancer cells. Cell. Signal. 2010, 22, 1369–1378. [Google Scholar] [CrossRef] [Green Version]
- Doggrell, S.A. RITA—A small-molecule anticancer drug that targets p53. Expert Opin. Investig. Drugs 2005, 14, 739–742. [Google Scholar] [CrossRef]
- Kaur, R.P.; Vasudeva, K.; Kumar, R.; Munshi, A. Role of p53 Gene in Breast Cancer: Focus on Mutation Spectrum and Therapeutic Strategies. Curr. Pharm. Des. 2018, 24, 3566–3575. [Google Scholar] [CrossRef]
- Gross, A.M.; Wolters, P.L.; Dombi, E.; Baldwin, A.; Whitcomb, P.; Fisher, M.J.; Weiss, B.; Kim, A.; Bornhorst, M.; Shah, A.C.; et al. Selumetinib in Children with Inoperable Plexiform Neurofibromas. N. Engl. J. Med. 2020, 382, 1430–1442. [Google Scholar] [CrossRef]
- Zhou, Y.; Lin, S.; Tseng, K.F.; Han, K.; Wang, Y.; Gan, Z.H.; Min, D.L.; Hu, H.Y. Selumetinib suppresses cell proliferation, migration and trigger apoptosis, G1 arrest in triple-negative breast cancer cells. BMC Cancer 2016, 16, 818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartholomeusz, C.; Xie, X.; Pitner, M.K.; Kondo, K.; Dadbin, A.; Lee, J.; Saso, H.; Smith, P.D.; Dalby, K.N.; Ueno, N.T. MEK Inhibitor Selumetinib (AZD6244; ARRY-142886) Prevents Lung Metastasis in a Triple-Negative Breast Cancer Xenograft Model. Mol. Cancer Ther. 2015, 14, 2773–2781. [Google Scholar] [CrossRef] [Green Version]
- Chargari, C.; Leteur, C.; Angevin, E.; Bashir, T.; Schoentjes, B.; Arts, J.; Janicot, M.; Bourhis, J.; Deutsch, E. Preclinical assessment of JNJ-26854165 (Serdemetan), a novel tryptamine compound with radiosensitizing activity in vitro and in tumor xenografts. Cancer Lett. 2011, 312, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Tabernero, J.; Dirix, L.; Schöffski, P.; Cervantes, A.; Lopez-Martin, J.A.; Capdevila, J.; van Beijsterveldt, L.; Platero, S.; Hall, B.; Yuan, Z.; et al. A phase I first-in-human pharmacokinetic and pharmacodynamic study of serdemetan in patients with advanced solid tumors. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2011, 17, 6313–6321. [Google Scholar] [CrossRef] [Green Version]
- Burstein, H.J.; Elias, A.D.; Rugo, H.S.; Cobleigh, M.A.; Wolff, A.C.; Eisenberg, P.D.; Lehman, M.; Adams, B.J.; Bello, C.L.; DePrimo, S.E.; et al. Phase II Study of Sunitinib Malate, an Oral Multitargeted Tyrosine Kinase Inhibitor, in Patients With Metastatic Breast Cancer Previously Treated With an Anthracycline and a Taxane. J. Clin. Oncol. 2008, 26, 1810–1816. [Google Scholar] [CrossRef]
- Liu, S.; Goldstein, R.H.; Scepansky, E.M.; Rosenblatt, M. Inhibition of Rho-Associated Kinase Signaling Prevents Breast Cancer Metastasis to Human Bone. Cancer Res. 2009, 69, 8742–8751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Landry, A.P.; Wang, J.Z.; Suppiah, S.; Zadeh, G. Multiplatform molecular analysis of vestibular schwannoma reveals two robust subgroups with distinct microenvironment. J. Neuro-Oncol. 2023, 161, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Zur, E. Low-dose Pilocarpine Spray to Treat Xerostomia. Int. J. Pharm. Compd. 2020, 24, 104–108. [Google Scholar] [PubMed]
- Trautner, E.M.; Gershon, S. Use of tremorine for screening anti-parkinsonian drugs. Nature 1959, 183, 1462–1463. [Google Scholar] [CrossRef] [PubMed]
- Grigoriadis, A.; Mackay, A.; Noel, E.; Wu, P.J.; Natrajan, R.; Frankum, J.; Reis-Filho, J.S.; Tutt, A. Molecular characterisation of cell line models for triple-negative breast cancers. BMC Genom. 2012, 13, 619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boichuk, S.; Galembikova, A.; Sitenkov, A.; Khusnutdinov, R.; Dunaev, P.; Valeeva, E.; Usolova, N. Establishment and characterization of a triple negative basal-like breast cancer cell line with multi-drug resistance. Oncol. Lett. 2017, 14, 5039–5045. [Google Scholar] [CrossRef] [Green Version]
- Chavez, K.J.; Garimella, S.V.; Lipkowitz, S. Triple negative breast cancer cell lines: One tool in the search for better treatment of triple negative breast cancer. Breast Dis. 2010, 32, 35–48. [Google Scholar] [CrossRef] [Green Version]
- Su, W.; Feng, B.; Hu, L.; Guo, X.; Yu, M. MUC3A promotes the progression of colorectal cancer through the PI3K/Akt/mTOR pathway. BMC Cancer 2022, 22, 602. [Google Scholar] [CrossRef]
- Kalvala, A.; Gao, L.; Aguila, B.; Dotts, K.; Rahman, M.; Nana-Sinkam, S.P.; Zhou, X.; Wang, Q.E.; Amann, J.; Otterson, G.A.; et al. Rad51C-ATXN7 fusion gene expression in colorectal tumors. Mol. Cancer 2016, 15, 47. [Google Scholar] [CrossRef] [Green Version]
- Ye, Q.; Falatovich, B.; Singh, S.; Ivanov, A.V.; Eubank, T.D.; Guo, N.L. A Multi-Omics Network of a Seven-Gene Prognostic Signature for Non-Small Cell Lung Cancer. Int. J. Mol. Sci. 2021, 23, 219. [Google Scholar] [CrossRef]
- Ye, Q.; Guo, N.L. Single B Cell Gene Co-Expression Networks Implicated in Prognosis, Proliferation, and Therapeutic Responses in Non-Small Cell Lung Cancer Bulk Tumors. Cancers 2022, 14, 3123. [Google Scholar] [CrossRef]
- Ye, Q.; Hickey, J.; Summers, K.; Falatovich, B.; Gencheva, M.; Eubank, T.D.; Ivanov, A.V.; Guo, N.L. Multi-Omics Immune Interaction Networks in Lung Cancer Tumorigenesis, Proliferation, and Survival. Int. J. Mol. Sci. 2022, 23, 14978. [Google Scholar] [CrossRef] [PubMed]








| Clinical Information | Sample Size (N) |
|---|---|
| Cancer Stage | |
| Ductal carcinoma in situ (DCIS) | 1 |
| 1 | 8 |
| 2 | 22 |
| 3 | 12 |
| 4 | 2 |
| Normal | 7 |
| Age | |
| <60 | 27 |
| 60–80 | 17 |
| >80 | 3 |
| Missing | 5 |
| Tumor Grade | |
| 1 | 3 |
| 2 | 13 |
| 3 | 25 |
| Missing | 11 |
| Histology | |
| Ductal adenocarcinoma | 1 |
| In situ carcinoma | 1 |
| Invasive ductal carcinoma | 35 |
| Invasive lobular carcinoma | 3 |
| Metaplastic carcinoma | 1 |
| Mucinous carcinoma | 1 |
| Normal tissue | 7 |
| Predominantly lobular | 1 |
| Spindle cell sarcoma | 1 |
| Missing | 1 |
| miRNA in Our Dataset | Fold Change (Unpaired) | Fold Change (Paired) | miRNA in TCGA | Logrank p-Value | Hazard Ratio (95% Confidence Interval) | TCGA Dataset | Potential Function Category |
|---|---|---|---|---|---|---|---|
| hsa-miR-100 | 0.316 | 0.295 | hsa-miR-100 | 0.0195 | 0.766 [0.613, 0.9576] | GA, n = 324 | tumor suppressor |
| hsa-miR-101 | 0.474 | 0.278 | hsa-miR-101-1 | 0.0021 | 0.691 [0.546, 0.8741] | HiSeq, n = 755 | tumor suppressor |
| hsa-miR-101-2 | 0.0278 | 0.805 [0.6626, 0.9769] | HiSeq, n = 755 | ||||
| hsa-miR-193a-3p | 2.070 | 2.244 | hsa-miR-193a | 0.0060 | 1.481 [1.1219, 1.9555] | HiSeq, n = 755 | oncogene |
| hsa-miR-204 | 0.241 | 0.148 | hsa-miR-204 | 0.0233 | 0.819 [0.6905, 0.9725] | HiSeq, n = 755 | tumor suppressor |
| hsa-miR-205 | 0.246 | 0.180 | hsa-miR-205 | 0.0335 | 0.871 [0.7667, 0.9904] | GA, n = 324 | tumor suppressor |
| hsa-miR-301b | 2.118 | 1.948 | hsa-miR-301b | 0.0100 | 1.410 [1.0837, 1.8342] | GA, n = 324 | oncogene |
| hsa-miR-30a | 0.406 | 0.324 | hsa-miR-30a | 0.0176 | 0.843 [0.7324, 0.9711] | HiSeq, n = 755 | tumor suppressor |
| hsa-miR-30a* | 0.351 | 0.250 | |||||
| hsa-miR-379 | 0.478 | 0.418 | hsa-miR-379 | 0.0480 | 0.797 [0.6369, 0.9981] | GA, n = 324 | tumor suppressor |
| hsa-miR-615-3p | 1.8673 | 2.531 | hsa-miR-615 | 0.0068 | 1.332 [1.0804, 1.6414] | GA, n = 324 | oncogene |
| hsa-miR-7 | 2.4657 | 3.994 | hsa-miR-7-2 | 0.0393 | 1.464 [1.0182, 2.1057] | GA, n = 324 | oncogene |
| hsa-miR-7-3 | 0.0106 | 1.543 [1.1036, 2.1582] | GA, n = 324 | ||||
| hsa-miR-720 | 4.2969 | 5.964 | hsa-miR-720 | 0.0179 | 2.867 [1.189, 6.9117] | HiSeq, n = 755 | oncogene |
| hsa-miR-99a | 0.2919 | 0.242 | hsa-miR-99a | 0.0128 | 0.744 [0.5901, 0.939] | GA, n = 324 | tumor suppressor |
| Drug | Pan-Sensitive Genes | Pan-Resistant Genes |
|---|---|---|
| 5-fluorouracil | GALM, HELLS, RBM14, DUOX1, RBM25, NOP56, DHX15, BAZ1B, DGCR8, CD276, SNX13, PTPRF, PHIP, TAOK1, SUZ12, PLEK, hsa-miR-1246, hsa-miR-617 | KIF3A, KAT2B, MYADM, DDX3Y, PTMS, EPHA7, EMC4, IRAK4, hsa-miR-33a, hsa-miR-1260 |
| alpelisib | DUOX1, PTPRF, ZBED6, DGCR8, TRABD, NOP56, RBM25, SUZ12, GALM, WDR82, CDS2, hsa-miR-617, hsa-miR-933 | KAT2B, EPHA7, BCL6, MTX3, IRGQ, TXNL1, IRAK4, GSK3A, APOD, KIF3A, MYADM, hsa-miR-1274a, hsa-miR-1280, hsa-miR-335, hsa-miR-376a, hsa-miR-720 |
| capecitabine | GSK3A, ZC3H4, IRGQ, BCL6, MGST2, LZTS2 | |
| carboplatin | WDR82 | |
| cisplatin | SNX13, GATB, PRDM2, MFSD9, PHIP, TBRG1, CBX3, RPRD2, RBM14, PTPRF, EGR1, DHX15, PDE3A, ATG4D, HERC6, DUOX1 | PTMS, PEG10, IRGQ, APOD, MTX3, PTEN, STX3, ZNF24, LZTS2, KIF3A, FRYL, MYADM, hsa-miR-301a, hsa-miR-497, hsa-miR-335, hsa-miR-1260 |
| cyclophosphamide | DHX15, RBM25, PHIP, DGCR8, PRDM2, RPRD2, PTPRF, hsa-miR-1246 | LZTS2, PTEN, FRYL, MYADM |
| docetaxel | PDE3A, TBRG1, BAZ1B, DHX15, TBRG1, RBM25, RPRD2, hsa-miR-1246, hsa-miR-142-5p | GSK3A, ZC3H4, STX3, STMN3, IRAK4, APOD, KAT2B, EPHA7, PEG10 |
| doxorubicin | CDS2, HERC6, NOP56, CD276, TRABD, MFSD9, PHIP, HELLS, GATB, BAZ1B, NKTR, RBM25, SUZ12, CBX3, CHD9, RBM14 | KIF3A, PTMS, IRGQ, GSK3A, TTC26, hsa-miR-301a |
| epirubicin | NKTR, RBM25, TRABD, GATB, LSAMP, DHX15, PLEK | PTEN, ZNF24, LZTS2, EMC4, ZC3H4, MGST2, hsa-miR-1260, hsa-miR-21, hsa-miR-301a |
| fulvestrant | BAZ1B, PRDM2, RBM25, CBX3, SUZ12, RBM14, HELLS, DHX15, NKTR, DGCR8, TAOK1, GATB, TRABD, LSAMP, RPRD2, CD276, hsa-miR-1976, hsa-miR-378, hsa-miR-766, hsa-miR-1246 | MGST2, IRGQ, PSG6, APOD, IRAK4, GSK3A, MYADM, hsa-miR-33a |
| gemcitabine | TBRG1, NOP56, MFSD9, RBM25, BAZ1B | APOD, PTMS, MAVS, ZC3H4, hsa-miR-301a |
| ixabepilone | DUOX1, TBRG1, ZBED6, RBM14, LSAMP, BAZ1B | LZTS2, STX3, PTEN, PEG10, KIF3A |
| lapatinib | DUOX1, HERC6, PLEK, PRDM2, CD276, hsa-miR-933 | ZNF24, TTC26, KAT2B, PTMS, IRGQ, TXNL1, IRAK4, KIF3A, MYADM, PSG6 |
| methotrexate | MFSD9, PHIP, RPRD2, CBX3, hsa-miR-1246 | MYADM, LZTS2, hsa-miR-497 |
| neratinib | SNX13, ZBED6, PRDM2, hsa-miR-378, hsa-miR-10b | TXNL1, MTX3, IRGQ, TTC26, EMC4, ZNF24, STMN3, hsa-miR-21 |
| olaparib | PTPRF, CHD9, GATB, PRDM2, BAZ1B, CBX3, PDE3A, PHIP, ZBED6, MFSD9, TBRG1, CD276, SNX13 | MTX3, FRYL, BCL6, KAT2B, LZTS2, APOD, DDX3Y, MYADM |
| paclitaxel | GALM, DHX15, LSAMP, SNX13 | MAVS, BCL6, ZC3H4, STX3, KAT2B, MTX3, KIF3A, STMN3, PEG10, MYADM, PTMS, hsa-miR-21, hsa-miR-497 |
| tucatinib | CDS2, ATG4D, SNX13, PLEK, DUOX1, GATB, CHD9, hsa-miR-205, hsa-miR-378, hsa-miR-933, hsa-miR-376c | MTX3, PTEN, ZNF24, APOD, GSK3A, hsa-miR-301a |
| vinorelbine | PRDM2, DGCR8, TRABD, EGR1, BAZ1B, CD276, PTPRF, GATB, ATG4D, TBRG1, TAOK1, WDR82, SUZ12, RBM14, NKTR, PDE3A, RPRD2 | MTX3, MAVS, TTC26, EPHA7, hsa-miR-21, hsa-miR-199b-5p |
| Drug | Sensitive Gene | Resistant Gene |
|---|---|---|
| Lestaurtinib | FHL1, FKBP7, LAMA4, LAMB3, LAMC1, MMP14 | ARRB1 |
| BRD-K12244279 | ACSL4, ASPH, CAV1, CAV2, EHD2, FMNL2, FSTL1, FYN, IFIT1, IFIT2, LDHB, SAMD9, UPP1, WLS | ENPP1 |
| PD198306 | ICAM1, IFIT3 | |
| Pilocarpine | SDC2 | |
| TG101348 | VLDLR | ANXA8, APOL1, FBLN1, ICAM1, MX2 |
| Tremorine | CRIP1, GPC4 | |
| U0126 | DKK3, FSD1, KYNU, LRP12 | |
| Y-27632 | C1R, CASP1, FOSL1, GSDMD | CLIC3, CPLX1, SYAP1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, Q.; Raese, R.A.; Luo, D.; Feng, J.; Xin, W.; Dong, C.; Qian, Y.; Guo, N.L. MicroRNA-Based Discovery of Biomarkers, Therapeutic Targets, and Repositioning Drugs for Breast Cancer. Cells 2023, 12, 1917. https://doi.org/10.3390/cells12141917
Ye Q, Raese RA, Luo D, Feng J, Xin W, Dong C, Qian Y, Guo NL. MicroRNA-Based Discovery of Biomarkers, Therapeutic Targets, and Repositioning Drugs for Breast Cancer. Cells. 2023; 12(14):1917. https://doi.org/10.3390/cells12141917
Chicago/Turabian StyleYe, Qing, Rebecca A. Raese, Dajie Luo, Juan Feng, Wenjun Xin, Chunlin Dong, Yong Qian, and Nancy Lan Guo. 2023. "MicroRNA-Based Discovery of Biomarkers, Therapeutic Targets, and Repositioning Drugs for Breast Cancer" Cells 12, no. 14: 1917. https://doi.org/10.3390/cells12141917