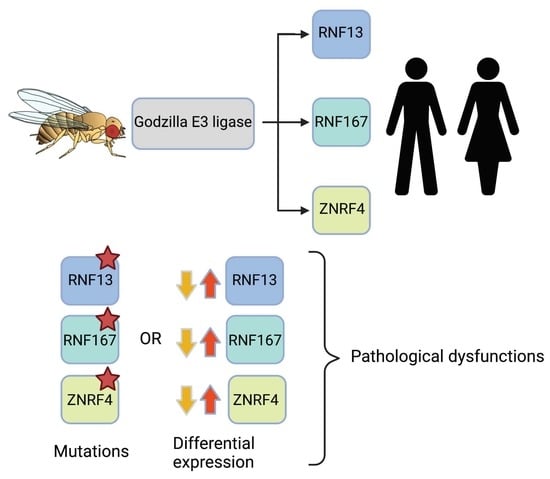
From Drosophila to Human: Biological Function of E3 Ligase Godzilla and Its Role in Disease
Abstract
:1. Introduction


2. Godzilla
2.1. Godzilla’s Substrate Regulates Recycling Endosomes
2.2. Wingless Transcytosis
3. RNF13
3.1. Protein Domains
3.1.1. PEST Domain
3.1.2. Nuclear Localization Signal
3.1.3. Di-Leucine Sorting Signal
3.2. Cellular Functions
3.2.1. Neurobiological Role
3.2.2. Cell Migration and Invasiveness
3.2.3. ER Stress
3.2.4. Myogenesis and Muscle Regeneration
3.3. Pathological Dysfunctions Associated with RNF13
3.3.1. Cancer
3.3.2. Atherosclerotic Plaques
3.3.3. Parkinson’s Disease
3.3.4. Developmental and Epileptic Encephalopathy 73
4. RNF167
4.1. Expression
4.2. Neurotransmission Modulator
4.3. Substrates Regulate Endosomal Trafficking and Lysosome Positioning
4.4. Pathological Dysfunctions Associated with RNF167
Cancer
5. ZNRF4
5.1. Calnexin
5.2. RIP2
6. Conclusions and Future Perspectives
| E3 Ligase | Substrate | Role of Ubiquitination | Biological Function | Pathological Dysfunction |
|---|---|---|---|---|
| Godzilla | Syb [33,45] | Not determined (N.D.) | Regulation of recycling endosome trafficking [33] | N.D. |
| N.D. | Wingless transcytosis [45] | N.D. | ||
| RNF13 | N.D. | N.D. | Ectopic expression promotes spontaneous growth of neurites in PC12 cells [60] | N.D. |
| N.D. | mRNA expression increases with dibuturyl-cAMP treatment in B35 cells [47] | N.D. | ||
| Snapin [62] | K29-linked poly-Ub | SNARE complex assembly [62] | Mice display deficit in learning [62] | |
| increased expression [62] | ||||
| N.D. | N.D. | Increased MMP-9 activity [49] | Cancer [49,66] | |
| N.D. | Regulation of GM-CSF concentration [66] | |||
| N.D. | Altered endolysosomal system [27,59] | Developmental and epileptic encephalopathy 73 [59] | ||
| Cancer [27] | ||||
| IRE1α | N.D. | Regulation of ER stress [48] | Developmental and epileptic encephalopathy 73 [74] | |
| (Interacts but not shown to be a substrate) [48] | Mouse model of Parkinson’s disease [73] | |||
| Atherosclerotic plaques [72] | ||||
| N.D. | N.D. | Regulation of muscle cell proliferation through regulation of IL-4 and IL-6 [65,68] | N.D. | |
| RNF167 | GluA2 [76,77] | Regulated surface expression [76] | Regulation of neuronal synaptic strength [76] | N.D. |
| Vamp3 [33] | N.D. | Regulation of recycling endosomes [33] | N.D. | |
| Arl8B [78] | Degradation via the proteasome-dependent pathway [78] | Lysosomal positioning [78] | Cancer [75,80,81] | |
| N.D. | N.D. | Lysosomal exocytosis [80] | ||
| CASTOR1 [81] | K29-linked polyUb chain | Inhibition of mTORC1 activation [81] | ||
| Degradation via the proteasome-dependent pathway [81] | ||||
| TSSC5 [75] | Degradation via the proteasome-dependent pathway [75] | Delays G1-to-S transition in HeLa cells [75] | ||
| ZNRF4 | N.D. | N.D. | Spermatogenesis in mice? [83] | N.D. |
| Calnexin [84] | Degradation via the proteasome-dependent pathway [84] | ER homeostasis [84] | N.D. | |
| RIP2 [85] | K48-linked poly-Ub chain | Negatively regulates NOD2 signaling [85] | N.D. | |
| Degradation via the proteasome-dependent pathway [85] |

| Gene | Post-Translational Modifications |
|---|---|
| RNF167 ID 26001 Position 17p13.2 | Ubiquitination: K97, K156, K210, K214, K223, K242, K266, K272 [98,99,100,101] Methylation: R277 [102] Phosphorylation: T288, S341, S345 [103] Glycosylation: N33, N79 [76] |
| RNF13 ID 11342 Position 3q25.1 | Ubiquitination: K107, K224, K225, K230, K232, K233, K252, K265, K273, K275, K282 [98,99,100,101,104,105,106] Phosphorylation: T309, S319, T380 [103,107] Glycosylation: N70, N75, N88 [49,108] |
| ZNRF4 ID 148066 Position 19p13.3 | Phosphorylation: T70 [109] Glycosylation: N107, N152, N229 [84] |
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AMPA | α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid |
| AMPAR | AMPA receptor |
| AMPAR-EPSC | Evoked excitatory postsynaptic AMPAR currents |
| AP | Adaptor protein |
| Arl8B | ADP-ribosylation factor-like protein 8B |
| ASK1 | Apoptosis signal-regulating kinase 1 |
| C-RZF | Chicken RING zinc finger |
| CASTOR1 | Cytosolic arginine sensor for mTORC1 subunit 1 |
| CFM | Chicken fetal myoblasts |
| CHO | Chinese hamster ovary |
| CTX | Cardiotoxin |
| DEE73 | Developmental and epileptic encephalopathy 73 |
| DN | Dominant-negative |
| DUB | Deubiquitinase |
| E | Glutamic acid |
| E1 | Ubiquitin-activating enzyme |
| E2 | Ubiquitin-conjugating enzyme |
| E3 | Ubiquitin ligase |
| EEA1 | Early endosome antigen 1 |
| EGFR | Epidermal growth factor receptor |
| ER | Endoplasmic reticulum |
| ER+ | Estrogen receptor positive |
| FZD6 | Frizzled-6 |
| GM-CSF | Granulocyte macrophage colony-stimulating factor |
| HECT | Homology to E6AP Carboxyl Terminus |
| HEK293T | Human embryonic kidney 293 |
| HER2+ | Human epidermal growth factor receptor 2 positive |
| HTS | High-throughput screening |
| IL-1β | Interleukin 1β |
| IL-4 | Interleukin-4 |
| IL-6 | Interleukin-6 |
| IL-8 | Interleukin-8 |
| INM | Inner nuclear membrane |
| IRE1α | Inositol-requiring enzyme 1α |
| JNK | c-Jun N-terminal kinase |
| Lamp1 | Lysosomal-associated membrane protein 1 |
| M6P | Mannose-6-Phosphate |
| MARCH | Membrane-associated RING-CH |
| MMP-9 | Matrix metallopeptidase 9 |
| MPTP | 1-methyl-4-phenyl-1,2.3.6-tetrahydropyridine |
| mTORC1 | Mammalian target of rapamycin complex 1 |
| NF-κB | Nuclear factor-κB |
| NLS | Nuclear localization signal |
| NOD2 | Nucleotide-binding oligomerization domain 2 |
| Ox-LDL | Oxidized low-density lipoprotein |
| P | Proline |
| PA-TM-RING | Protease-associated transmembrane RING |
| PA | Protease-associated |
| PDAC | Pancreatic ductal adenocarcinoma |
| PI | Propidium iodide |
| PKC | Protein kinase C |
| PMA | Phorbol 12-myristate 13-acetate |
| PTM | Post-translational modification |
| RBR | RING-Between-RING |
| RING | Really Interesting Novel Gene |
| RIP2 | Receptor-interacting protein 2 |
| RNF13 | Ring Finger Protein 13 |
| RNF167 | Ring Finger Protein 167 |
| S | Serine |
| shRNF13 | Short hairpin RNA specific for RNF13 |
| siRNA | Small interfering RNA |
| SLO | Streptolysin-O |
| SNAP-25 | Synaptosome associated protein 25 |
| Snapin | SNARE-associated protein |
| SNARE | Soluble N-ethylmaleimide-sensitive-factor Attachment protein Receptor |
| SNV | Single-nucleotide variations |
| SP | Signal peptide |
| Sperizin | Spermatic-specific ring zinc finger |
| STS | Staurosporine |
| Syb | Synaptobrevin |
| T | Threonine |
| TA | Tibialis anterior |
| TH-IR | Tyrosine hydroxylase-immunoreactive |
| TM | Transmembrane |
| TNF-α | Tumor necrosis factor α |
| TNF | Tumor necrosis factor |
| TRAF2 | TNF receptor-associated factor 2 |
| TRIM | Tripartite motif-containing |
| TSSC5 | Tumor-Suppressing Subchromosomal Transferable Fragment cDNA |
| Ub | Ubiquitin |
| Vamp3 | Vesicle-associated membrane protein 3 |
| WT | Wild type |
| XBP1 | X-Box Binding Protein 1 |
| ZNRF4 | Zinc and Ring Finger 4 |
References
- Mann, M.; Jensen, O.N. Proteomic analysis of post-translational modifications. Nat. Biotechnol. 2003, 21, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Qian, C.; Cao, X. Post-Translational Modification Control of Innate Immunity. Immunity 2016, 45, 15–30. [Google Scholar] [CrossRef] [PubMed]
- Komander, D.; Rape, M. The ubiquitin code. Annu. Rev. Biochem. 2012, 81, 203–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hicke, L.; Dunn, R. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu. Rev. Cell Dev. Biol. 2003, 19, 141–172. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Rape, M. Building ubiquitin chains: E2 enzymes at work. Nat. Rev. Mol. Cell Biol. 2009, 10, 755–764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, Q.; Huang, T.; Zhang, L.; Zhou, Y.; Luo, H.; Xu, H.; Wang, X. Dysregulation of Ubiquitin-Proteasome System in Neurodegenerative Diseases. Front. Aging Neurosci. 2016, 8, 303. [Google Scholar] [CrossRef] [PubMed]
- Swatek, K.N.; Komander, D. Ubiquitin modifications. Cell Res. 2016, 26, 399–422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Wijk, S.J.L.; Fulda, S.; Dikic, I.; Heilemann, M. Visualizing ubiquitination in mammalian cells. EMBO Rep. 2019, 20, e46520. [Google Scholar] [CrossRef]
- Behrends, C.; Harper, J.W. Constructing and decoding unconventional ubiquitin chains. Nat. Struct. Mol. Biol. 2011, 18, 520–528. [Google Scholar] [CrossRef]
- Rahighi, S.; Ikeda, F.; Kawasaki, M.; Akutsu, M.; Suzuki, N.; Kato, R.; Kensche, T.; Uejima, T.; Bloor, S.; Komander, D.; et al. Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell 2009, 136, 1098–1109. [Google Scholar] [CrossRef] [Green Version]
- Noad, J.; von der Malsburg, A.; Pathe, C.; Michel, M.A.; Komander, D.; Randow, F. LUBAC-synthesized linear ubiquitin chains restrict cytosol-invading bacteria by activating autophagy and NF-κB. Nat. Microbiol. 2017, 2, 17063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Wijk, S.J.L.; Fricke, F.; Herhaus, L.; Gupta, J.; Hötte, K.; Pampaloni, F.; Grumati, P.; Kaulich, M.; Sou, Y.S.; Komatsu, M.; et al. Linear ubiquitination of cytosolic Salmonella Typhimurium activates NF-κB and restricts bacterial proliferation. Nat. Microbiol. 2017, 2, 17066. [Google Scholar] [CrossRef] [PubMed]
- Tracz, M.; Bialek, W. Beyond K48 and K63: Non-canonical protein ubiquitination. Cell. Mol. Biol. Lett. 2021, 26, 1. [Google Scholar] [CrossRef] [PubMed]
- van Huizen, M.; Kikkert, M. The Role of Atypical Ubiquitin Chains in the Regulation of the Antiviral Innate Immune Response. Front. Cell Dev. Biol. 2020, 7, 392. [Google Scholar] [CrossRef]
- Sigismund, S.; Polo, S.; Di Fiore, P.P. Signaling through monoubiquitination. Curr. Top Microbiol. Immunol. 2004, 286, 149–185. [Google Scholar] [CrossRef]
- Sun, L.; Chen, Z.J. The novel functions of ubiquitination in signaling. Curr. Opin. Cell Biol. 2004, 16, 119–126. [Google Scholar] [CrossRef]
- Kravtsova-Ivantsiv, Y.; Sommer, T.; Ciechanover, A. The lysine48-based polyubiquitin chain proteasomal signal: Not a single child anymore. Angew. Chem. Int. Ed. Engl. 2013, 52, 192–198. [Google Scholar] [CrossRef]
- Chen, Z.J. Ubiquitin signalling in the NF-kappaB pathway. Nat. Cell Biol. 2005, 7, 758–765. [Google Scholar] [CrossRef] [Green Version]
- Suresh, B.; Lee, J.; Kim, H.; Ramakrishna, S. Regulation of pluripotency and differentiation by deubiquitinating enzymes. Cell Death Differ. 2016, 23, 1257–1264. [Google Scholar] [CrossRef] [Green Version]
- Chastagner, P.; Israël, A.; Brou, C. Itch/AIP4 mediates Deltex degradation through the formation of K29-linked polyubiquitin chains. EMBO Rep. 2006, 7, 1147–1153. [Google Scholar] [CrossRef]
- Matsumoto, M.L.; Wickliffe, K.E.; Dong, K.C.; Yu, C.; Bosanac, I.; Bustos, D.; Phu, L.; Kirkpatrick, D.S.; Hymowitz, S.G.; Rape, M.; et al. K11-linked polyubiquitination in cell cycle control revealed by a K11 linkage-specific antibody. Mol. Cell 2010, 39, 477–484. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.H.; Beaudet, A.L. Human disorders of ubiquitination and proteasomal degradation. Curr. Opin. Pediatr. 2004, 16, 419–426. [Google Scholar] [CrossRef]
- Spratt, D.E.; Walden, H.; Shaw, G.S. RBR E3 ubiquitin ligases: New structures, new insights, new questions. Biochem. J. 2014, 458, 421–437. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Bengtson, M.H.; Ulbrich, A.; Matsuda, A.; Reddy, V.A.; Orth, A.; Chanda, S.K.; Batalov, S.; Joazeiro, C.A.P. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle’s dynamics and signaling. PLoS ONE 2008, 3, e1487. [Google Scholar] [CrossRef] [PubMed]
- Metzger, M.B.; Hristova, V.A.; Weissman, A.M. HECT and RING finger families of E3 ubiquitin ligases at a glance. J. Cell Sci. 2012, 125, 531–537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakamura, N. The Role of the Transmembrane RING Finger Proteins in Cellular and Organelle Function. Membranes 2011, 1, 354–393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Dijk, J.R.; Yamazaki, Y.; Palmer, R.H. Tumour-associated mutations of PA-TM-RING ubiquitin ligases RNF167/RNF13 identify the PA domain as a determinant for endosomal localization. Biochem. J. 2014, 459, 27–36. [Google Scholar] [CrossRef]
- Nakai, K.; Imai, K. Prediction of Protein Localization. In Encyclopedia of Bioinformatics and Computational Biology; Ranganathan, S., Gribskov, M., Nakai, K., Schönbach, C., Eds.; Academic Press: Oxford, UK, 2019; pp. 53–59. [Google Scholar] [CrossRef]
- Mahon, P.; Bateman, A. The PA domain: A protease-associated domain. Protein Sci. 2000, 9, 1930–1934. [Google Scholar] [CrossRef] [Green Version]
- Luo, X.; Hofmann, K. The protease-associated domain: A homology domain associated with multiple classes of proteases. Trends Biochem. Sci. 2001, 26, 147–148. [Google Scholar] [CrossRef]
- Wayne Albers, R.R.W. Chapter 2-Cell Membrane Structures and Functions. In Basic Neurochemistry, 8th ed.; Brady, S.T., Siegel, G.J., Albers, R.W., Price, D.L., Eds.; Academic Press: New York, NY, USA, 2012; pp. 26–39. [Google Scholar] [CrossRef]
- Jenn, R.C. A Systematic Analysis of Human Transmembrane E3-RING Proteins. Doctoral Dissertation, University of Liverpool, Liverpool, UK, 2011. [Google Scholar]
- Yamazaki, Y.; Schönherr, C.; Varshney, G.K.; Dogru, M.; Hallberg, B.; Palmer, R.H. Goliath family E3 ligases regulate the recycling endosome pathway via VAMP3 ubiquitylation. EMBO J. 2013, 32, 524–537. [Google Scholar] [CrossRef] [Green Version]
- Côté, S.; Preiss, A.; Haller, J.; Schuh, R.; Kienlin, A.; Seifert, E.; Jäckle, H. The gooseberry-zipper region of Drosophila: Five genes encode different spatially restricted transcripts in the embryo. EMBO J. 1987, 6, 2793–2801. [Google Scholar] [CrossRef] [PubMed]
- Bouchard, M.L.; Côté, S. The Drosophila melanogaster developmental gene g1 encodes a variant zinc-finger-motif protein. Gene 1993, 125, 205–209. [Google Scholar] [CrossRef]
- Artero, R.; Furlong, E.E.; Beckett, K.; Scott, M.P.; Baylies, M. Notch and Ras signaling pathway effector genes expressed in fusion competent and founder cells during Drosophila myogenesis. Development 2003, 130, 6257–6272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, J.; Wen, L.; Gao, X.; Jin, C.; Xue, Y.; Yao, X. DOG 1.0: Illustrator of protein domain structures. Cell Res. 2009, 19, 271–273. [Google Scholar] [CrossRef] [PubMed]
- Anandasabapathy, N.; Ford, G.S.; Bloom, D.; Holness, C.; Paragas, V.; Seroogy, C.; Skrenta, H.; Hollenhorst, M.; Fathman, C.G.; Soares, L. GRAIL: An E3 ubiquitin ligase that inhibits cytokine gene transcription is expressed in anergic CD4+ T cells. Immunity 2003, 18, 535–547. [Google Scholar] [CrossRef] [Green Version]
- Soares, L.; Seroogy, C.; Skrenta, H.; Anandasabapathy, N.; Lovelace, P.; Chung, C.D.; Engleman, E.; Fathman, C.G. Two isoforms of otubain 1 regulate T cell anergy via GRAIL. Nat. Immunol. 2004, 5, 45–54. [Google Scholar] [CrossRef]
- Heissmeyer, V.; Macián, F.; Im, S.H.; Varma, R.; Feske, S.; Venuprasad, K.; Gu, H.; Liu, Y.C.; Dustin, M.L.; Rao, A. Calcineurin imposes T cell unresponsiveness through targeted proteolysis of signaling proteins. Nat. Immunol. 2004, 5, 255–265. [Google Scholar] [CrossRef]
- Adams, M.D.; Celniker, S.E.; Holt, R.A.; Evans, C.A.; Gocayne, J.D.; Amanatides, P.G.; Scherer, S.E.; Li, P.W.; Hoskins, R.A.; Galle, R.F.; et al. The genome sequence of Drosophila melanogaster. Science 2000, 287, 2185–2195. [Google Scholar] [CrossRef] [Green Version]
- McMahon, H.T.; Ushkaryov, Y.A.; Edelmann, L.; Link, E.; Binz, T.; Niemann, H.; Jahn, R.; Südhof, T.C. Cellubrevin is a ubiquitous tetanus-toxin substrate homologous to a putative synaptic vesicle fusion protein. Nature 1993, 364, 346–349. [Google Scholar] [CrossRef]
- Galli, T.; Chilcote, T.; Mundigl, O.; Binz, T.; Niemann, H.; De Camilli, P. Tetanus toxin-mediated cleavage of cellubrevin impairs exocytosis of transferrin receptor-containing vesicles in CHO cells. J. Cell Biol. 1994, 125, 1015–1024. [Google Scholar] [CrossRef]
- Tuma, P.L.; Hubbard, A.L. Transcytosis: Crossing Cellular Barriers. Physiol. Rev. 2003, 83, 871–932. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, Y.; Palmer, L.; Alexandre, C.; Kakugawa, S.; Beckett, K.; Gaugue, I.; Palmer, R.H.; Vincent, J.P. Godzilla-dependent transcytosis promotes Wingless signalling in Drosophila wing imaginal discs. Nat. Cell. Biol. 2016, 18, 451–457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tranque, P.; Crossin, K.L.; Cirelli, C.; Edelman, G.M.; Mauro, V.P. Identification and characterization of a RING zinc finger gene (C-RZF) expressed in chicken embryo cells. Proc. Natl. Acad. Sci. USA 1996, 93, 3105–3109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bocock, J.P.; Carmicle, S.; Chhotani, S.; Ruffolo, M.R.; Chu, H.; Erickson, A.H. The PA-TM-RING protein RING finger protein 13 is an endosomal integral membrane E3 ubiquitin ligase whose RING finger domain is released to the cytoplasm by proteolysis. FEBS J. 2009, 276, 1860–1877. [Google Scholar] [CrossRef] [PubMed]
- Arshad, M.; Ye, Z.; Gu, X.; Wong, C.K.; Liu, Y.; Li, D.; Zhou, L.; Zhang, Y.; Bay, W.P.; Yu, V.C.; et al. RNF13, a RING finger protein, mediates endoplasmic reticulum stress-induced apoptosis through the inositol-requiring enzyme (IRE1α)/c-Jun NH2-terminal kinase pathway. J. Biol. Chem. 2013, 288, 8726–8736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Q.; Meng, Y.; Zhang, L.; Chen, J.; Zhu, D. RNF13: A novel RING-type ubiquitin ligase over-expressed in pancreatic cancer. Cell Res. 2009, 19, 348–357. [Google Scholar] [CrossRef]
- Bocock, J.P.; Carmicle, S.; Madamba, E.; Erickson, A.H. Nuclear targeting of an endosomal E3 ubiquitin ligase. Traffic 2010, 11, 756–766. [Google Scholar] [CrossRef]
- Rogers, S.; Wells, R.; Rechsteiner, M. Amino Acid Sequences Common to Rapidly Degraded Proteins: The PEST Hypothesis. Science 1986, 234, 364–368. [Google Scholar] [CrossRef]
- Bonifacino, J.S.; Traub, L.M. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 2003, 72, 395–447. [Google Scholar] [CrossRef] [Green Version]
- Boehm, M.; Bonifacino, J.S. Adaptins: The final recount. Mol. Biol. Cell 2001, 12, 2907–2920. [Google Scholar] [CrossRef]
- Huttlin, E.L.; Ting, L.; Bruckner, R.J.; Gebreab, F.; Gygi, M.P.; Szpyt, J.; Tam, S.; Zarraga, G.; Colby, G.; Baltier, K.; et al. The BioPlex Network: A Systematic Exploration of the Human Interactome. Cell 2015, 162, 425–440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huttlin, E.L.; Bruckner, R.J.; Paulo, J.A.; Cannon, J.R.; Ting, L.; Baltier, K.; Colby, G.; Gebreab, F.; Gygi, M.P.; Parzen, H.; et al. Architecture of the human interactome defines protein communities and disease networks. Nature 2017, 545, 505–509. [Google Scholar] [CrossRef] [PubMed]
- Huttlin, E.L.; Bruckner, R.J.; Navarrete-Perea, J.; Cannon, J.R.; Baltier, K.; Gebreab, F.; Gygi, M.P.; Thornock, A.; Zarraga, G.; Tam, S.; et al. Dual proteome-scale networks reveal cell-specific remodeling of the human interactome. Cell 2021, 184, 3022–3040.e28. [Google Scholar] [CrossRef] [PubMed]
- Sanger, A.; Hirst, J.; Davies, A.K.; Robinson, M.S. Adaptor protein complexes and disease at a glance. J. Cell Sci. 2019, 132, jcs222992. [Google Scholar] [CrossRef]
- Park, S.Y.; Guo, X. Adaptor protein complexes and intracellular transport. Biosci. Rep. 2014, 34, e00123. [Google Scholar] [CrossRef]
- Cabana, V.C.; Bouchard, A.Y.; Sénécal, A.M.; Ghilarducci, K.; Kourrich, S.; Cappadocia, L.; Lussier, M.P. RNF13 Dileucine Motif Variants L311S and L312P Interfere with Endosomal Localization and AP-3 Complex Association. Cells 2021, 10, 3063. [Google Scholar] [CrossRef]
- Saito, S.; Honma, K.; Kita-Matsuo, H.; Ochiya, T.; Kato, K. Gene expression profiling of cerebellar development with high-throughput functional analysis. Physiol. Genom. 2005, 22, 8–13. [Google Scholar] [CrossRef] [Green Version]
- Kaneko, M.; Iwase, I.; Yamasaki, Y.; Takai, T.; Wu, Y.; Kanemoto, S.; Matsuhisa, K.; Asada, R.; Okuma, Y.; Watanabe, T.; et al. Genome-wide identification and gene expression profiling of ubiquitin ligases for endoplasmic reticulum protein degradation. Sci. Rep. 2016, 6, 30955. [Google Scholar] [CrossRef]
- Zhang, Q.; Li, Y.; Zhang, L.; Yang, N.; Meng, J.; Zuo, P.; Zhang, Y.; Chen, J.; Wang, L.; Gao, X.; et al. E3 ubiquitin ligase RNF13 involves spatial learning and assembly of the SNARE complex. Cell Mol. Life Sci. 2013, 70, 153–165. [Google Scholar] [CrossRef]
- Al-Hakim, A.K.; Zagorska, A.; Chapman, L.; Deak, M.; Peggie, M.; Alessi, D.R. Control of AMPK-related kinases by USP9X and atypical Lys(29)/Lys(33)-linked polyubiquitin chains. Biochem. J. 2008, 411, 249–260. [Google Scholar] [CrossRef] [Green Version]
- Jin, X.; Cheng, H.; Chen, J.; Zhu, D. RNF13: An emerging RING finger ubiquitin ligase important in cell proliferation. FEBS J. 2011, 278, 78–84. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Wang, K.; Zhang, Y.; Meng, J.; Yu, F.; Chen, Y.; Zhu, D. The myostatin-induced E3 ubiquitin ligase RNF13 negatively regulates the proliferation of chicken myoblasts. FEBS J. 2010, 277, 466–476. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Wang, A.; Meng, J.; Zhang, Y.; Zhu, D. Enhanced metastasis in RNF13 knockout mice is mediated by a reduction in GM-CSF levels. Protein Cell 2015, 6, 746–756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arshad, A.; Gu, X.; Arshad, M. RNF13 protein regulates endoplasmic reticulum stress induced apoptosis in dopaminergic SH-SY5Y cells by enhancing IRE1α stability. J. Recept Signal Transduct. Res. 2014, 34, 119–124. [Google Scholar] [CrossRef]
- Meng, J.; Zou, X.; Wu, R.; Zhong, R.; Zhu, D.; Zhang, Y. Accelerated regeneration of the skeletal muscle in RNF13-knockout mice is mediated by macrophage-secreted IL-4/IL-6. Protein Cell 2014, 5, 235–247. [Google Scholar] [CrossRef] [Green Version]
- Ness, C.; Katta, K.; Garred, Ø.; Kumar, T.; Olstad, O.K.; Petrovski, G.; Moe, M.C.; Noer, A. Integrated differential DNA methylation and gene expression of formalin-fixed paraffin-embedded uveal melanoma specimens identifies genes associated with early metastasis and poor prognosis. Exp. Eye Res. 2020, 203, 108426. [Google Scholar] [CrossRef]
- Zhang, K.; Qin, X.; Zhou, X.; Zhou, J.; Wen, P.; Chen, S.; Wu, M.; Wu, Y.; Zhuang, J. Analysis of genes and underlying mechanisms involved in foam cells formation and atherosclerosis development. PeerJ 2020, 8, e10336. [Google Scholar] [CrossRef]
- Bazan-Socha, S.; Buregwa-Czuma, S.; Jakiela, B.; Zareba, L.; Zawlik, I.; Myszka, A.; Soja, J.; Okon, K.; Zarychta, J.; Kozlik, P.; et al. Reticular Basement Membrane Thickness Is Associated with Growth- and Fibrosis-Promoting Airway Transcriptome Profile-Study in Asthma Patients. Int. J. Mol. Sci. 2021, 22, 998. [Google Scholar] [CrossRef]
- Huang, D.; Liu, Y.; Gao, L.; Wei, X.; Xu, Y.; Cai, R.; Su, Q. MiR-32-3p Regulates Myocardial Injury Induced by Microembolism and Microvascular Obstruction by Targeting RNF13 to Regulate the Stability of Atherosclerotic Plaques. J. Cardiovasc. Transl. Res. 2021. [Google Scholar] [CrossRef]
- Ji, M.; Niu, S.; Guo, J.; Mi, H.; Jiang, P. Silencing RNF13 Alleviates Parkinson’s Disease-like Problems in Mouse Models by Regulating the Endoplasmic Reticulum Stress-Mediated IRE1α-TRAF2-ASK1-JNK Pathway. J. Mol. Neurosci. 2020, 70, 1977–1986. [Google Scholar] [CrossRef]
- Edvardson, S.; Nicolae, C.M.; Noh, G.J.; Burton, J.E.; Punzi, G.; Shaag, A.; Bischetsrieder, J.; De Grassi, A.; Pierri, C.L.; Elpeleg, O.; et al. Heterozygous RNF13 Gain-of-Function Variants Are Associated with Congenital Microcephaly, Epileptic Encephalopathy, Blindness, and Failure to Thrive. Am. J. Hum. Genet. 2019, 104, 179–185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamada, H.Y.; Gorbsky, G.J. Tumor suppressor candidate TSSC5 is regulated by UbcH6 and a novel ubiquitin ligase RING105. Oncogene 2006, 25, 1330–1339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lussier, M.P.; Herring, B.E.; Nasu-Nishimura, Y.; Neutzner, A.; Karbowski, M.; Youle, R.J.; Nicoll, R.A.; Roche, K.W. Ubiquitin ligase RNF167 regulates AMPA receptor-mediated synaptic transmission. Proc. Natl. Acad. Sci. USA 2012, 109, 19426–19431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghilarducci, K.; Cabana, V.C.; Desroches, C.; Chabi, K.; Bourgault, S.; Cappadocia, L.; Lussier, M.P. Functional interaction of ubiquitin ligase RNF167 with UBE2D1 and UBE2N promotes ubiquitination of AMPA receptor. FEBS J. 2021, 288, 4849–4868. [Google Scholar] [CrossRef] [PubMed]
- Deshar, R.; Moon, S.; Yoo, W.; Cho, E.B.; Yoon, S.K.; Yoon, J.B. RNF167 targets Arl8B for degradation to regulate lysosome positioning and endocytic trafficking. FEBS J. 2016, 283, 4583–4599. [Google Scholar] [CrossRef] [PubMed]
- Widagdo, J.; Chai, Y.J.; Ridder, M.C.; Chau, Y.Q.; Johnson, R.C.; Sah, P.; Huganir, R.L.; Anggono, V. Activity-Dependent Ubiquitination of GluA1 and GluA2 Regulates AMPA Receptor Intracellular Sorting and Degradation. Cell Rep. 2015, 10, 783–795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nair, S.V.; Narendradev, N.D.; Nambiar, R.P.; Kumar, R.; Srinivasula, S.M. Naturally occurring and tumor-associated variants of RNF167 promote lysosomal exocytosis and plasma membrane resealing. J. Cell Sci. 2020, 133, jcs239335. [Google Scholar] [CrossRef]
- Li, T.; Wang, X.; Ju, E.; da Silva, S.R.; Chen, L.; Zhang, X.; Wei, S.; Gao, S.J. RNF167 activates mTORC1 and promotes tumorigenesis by targeting CASTOR1 for ubiquitination and degradation. Nat. Commun. 2021, 12, 1055. [Google Scholar] [CrossRef]
- Fujii, T.; Tamura, K.; Copeland, N.G.; Gilbert, D.J.; Jenkins, N.A.; Yomogida, K.; Tanaka, H.; Nishimune, Y.; Nojima, H.; Abiko, Y. Sperizin is a murine RING zinc-finger protein specifically expressed in Haploid germ cells. Genomics 1999, 57, 94–101. [Google Scholar] [CrossRef]
- Glozak, M.A.; Li, Y.; Reuille, R.; Kim, K.H.; Vo, M.-N.; Rogers, M.B. Trapping and Characterization of Novel Retinoid Response Elements. Mol. Endocrinol. 2003, 17, 27–41. [Google Scholar] [CrossRef] [Green Version]
- Neutzner, A.; Neutzner, M.; Benischke, A.S.; Ryu, S.W.; Frank, S.; Youle, R.J.; Karbowski, M. A systematic search for endoplasmic reticulum (ER) membrane-associated RING finger proteins identifies Nixin/ZNRF4 as a regulator of calnexin stability and ER homeostasis. J. Biol. Chem. 2011, 286, 8633–8643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bist, P.; Cheong, W.S.; Ng, A.; Dikshit, N.; Kim, B.-H.; Pulloor, N.K.; Khameneh, H.J.; Hedl, M.; Shenoy, A.R.; Balamuralidhar, V.; et al. E3 Ubiquitin ligase ZNRF4 negatively regulates NOD2 signalling and induces tolerance to MDP. Nat. Commun. 2017, 8, 15865. [Google Scholar] [CrossRef] [PubMed]
- Attene-Ramos, M.S.; Austin, C.P.; Xia, M. High Throughput Screening. In Encyclopedia of Toxicology, 3rd ed.; Wexler, P., Ed.; Academic Press: Oxford, UK, 2014; pp. 916–917. [Google Scholar] [CrossRef]
- van Wijk, S.J.L.; de Vries, S.J.; Kemmeren, P.; Huang, A.; Boelens, R.; Bonvin, A.M.J.J.; Timmers, H.T.M. A comprehensive framework of E2–RING E3 interactions of the human ubiquitin–proteasome system. Mol. Syst. Biol. 2009, 5, 295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Guo, N.; Nathans, J. The role of Frizzled3 and Frizzled6 in neural tube closure and in the planar polarity of inner-ear sensory hair cells. J. Neurosci. 2006, 26, 2147–2156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stuebner, S.; Faus-Kessler, T.; Fischer, T.; Wurst, W.; Prakash, N. Fzd3 and Fzd6 deficiency results in a severe midbrain morphogenesis defect. Dev. Dyn. 2010, 239, 246–260. [Google Scholar] [CrossRef] [PubMed]
- Blandin, G.; Marchand, S.; Charton, K.; Danièle, N.; Gicquel, E.; Boucheteil, J.-B.; Bentaib, A.; Barrault, L.; Stockholm, D.; Bartoli, M.; et al. A human skeletal muscle interactome centered on proteins involved in muscular dystrophies: LGMD interactome. Skelet. Muscle 2013, 3, 3. [Google Scholar] [CrossRef] [Green Version]
- Antoniali, G.; Serra, F.; Lirussi, L.; Tanaka, M.; D’Ambrosio, C.; Zhang, S.; Radovic, S.; Dalla, E.; Ciani, Y.; Scaloni, A.; et al. Mammalian APE1 controls miRNA processing and its interactome is linked to cancer RNA metabolism. Nat. Commun. 2017, 8, 797. [Google Scholar] [CrossRef] [Green Version]
- Ewing, R.M.; Chu, P.; Elisma, F.; Li, H.; Taylor, P.; Climie, S.; McBroom-Cerajewski, L.; Robinson, M.D.; O’Connor, L.; Li, M.; et al. Large-scale mapping of human protein–protein interactions by mass spectrometry. Mol. Syst. Biol. 2007, 3, 89. [Google Scholar] [CrossRef]
- Hein, M.Y.; Hubner, N.C.; Poser, I.; Cox, J.; Nagaraj, N.; Toyoda, Y.; Gak, I.A.; Weisswange, I.; Mansfeld, J.; Buchholz, F.; et al. A Human Interactome in Three Quantitative Dimensions Organized by Stoichiometries and Abundances. Cell 2015, 163, 712–723. [Google Scholar] [CrossRef] [Green Version]
- Castello, A.; Fischer, B.; Eichelbaum, K.; Horos, R.; Beckmann, B.M.; Strein, C.; Davey, N.E.; Humphreys, D.T.; Preiss, T.; Steinmetz, L.M.; et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 2012, 149, 1393–1406. [Google Scholar] [CrossRef] [Green Version]
- van den Boomen, D.J.H.; Sienkiewicz, A.; Berlin, I.; Jongsma, M.L.M.; van Elsland, D.M.; Luzio, J.P.; Neefjes, J.J.C.; Lehner, P.J. A trimeric Rab7 GEF controls NPC1-dependent lysosomal cholesterol export. Nat. Commun. 2020, 11, 5559. [Google Scholar] [CrossRef] [PubMed]
- Luck, K.; Kim, D.-K.; Lambourne, L.; Spirohn, K.; Begg, B.E.; Bian, W.; Brignall, R.; Cafarelli, T.; Campos-Laborie, F.J.; Charloteaux, B.; et al. A reference map of the human binary protein interactome. Nature 2020, 580, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Fenech, E.J.; Lari, F.; Charles, P.D.; Fischer, R.; Laétitia-Thézénas, M.; Bagola, K.; Paton, A.W.; Paton, J.C.; Gyrd-Hansen, M.; Kessler, B.M.; et al. Interaction mapping of endoplasmic reticulum ubiquitin ligases identifies modulators of innate immune signalling. Elife 2020, 9, e57306. [Google Scholar] [CrossRef] [PubMed]
- Udeshi, N.D.; Svinkina, T.; Mertins, P.; Kuhn, E.; Mani, D.R.; Qiao, J.W.; Carr, S.A. Refined preparation and use of anti-diglycine remnant (K-ε-GG) antibody enables routine quantification of 10,000s of ubiquitination sites in single proteomics experiments. Mol. Cell Proteom. 2013, 12, 825–831. [Google Scholar] [CrossRef] [Green Version]
- Akimov, V.; Barrio-Hernandez, I.; Hansen, S.V.F.; Hallenborg, P.; Pedersen, A.K.; Bekker-Jensen, D.B.; Puglia, M.; Christensen, S.D.K.; Vanselow, J.T.; Nielsen, M.M.; et al. UbiSite approach for comprehensive mapping of lysine and N-terminal ubiquitination sites. Nat. Struct. Mol. Biol. 2018, 25, 631–640. [Google Scholar] [CrossRef]
- Beltrao, P.; Albanèse, V.; Kenner, L.R.; Swaney, D.L.; Burlingame, A.; Villén, J.; Lim, W.A.; Fraser, J.S.; Frydman, J.; Krogan, N.J. Systematic functional prioritization of protein posttranslational modifications. Cell 2012, 150, 413–425. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Wang, H.; Yang, Y.; Zhao, T.; Zhang, Z.; Tian, Y.; Shi, Z.; Peng, X.; Li, F.; Feng, Y.; et al. Integrative Analysis of Proteome and Ubiquitylome Reveals Unique Features of Lysosomal and Endocytic Pathways in Gefitinib-Resistant Non-Small Cell Lung Cancer Cells. Proteomics 2018, 18, e1700388. [Google Scholar] [CrossRef] [Green Version]
- Larsen, S.C.; Sylvestersen, K.B.; Mund, A.; Lyon, D.; Mullari, M.; Madsen, M.V.; Daniel, J.A.; Jensen, L.J.; Nielsen, M.L. Proteome-wide analysis of arginine monomethylation reveals widespread occurrence in human cells. Sci. Signal. 2016, 9, rs9. [Google Scholar] [CrossRef]
- Mertins, P.; Mani, D.R.; Ruggles, K.V.; Gillette, M.A.; Clauser, K.R.; Wang, P.; Wang, X.; Qiao, J.W.; Cao, S.; Petralia, F.; et al. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature 2016, 534, 55–62. [Google Scholar] [CrossRef] [Green Version]
- Stukalov, A.; Girault, V.; Grass, V.; Karayel, O.; Bergant, V.; Urban, C.; Haas, D.A.; Huang, Y.; Oubraham, L.; Wang, A.; et al. Multilevel proteomics reveals host perturbations by SARS-CoV-2 and SARS-CoV. Nature 2021, 594, 246–252. [Google Scholar] [CrossRef]
- Zhang, H.; Zheng, H.; Zhu, J.; Dong, Q.; Wang, J.; Fan, H.; Chen, Y.; Zhang, X.; Han, X.; Li, Q.; et al. Ubiquitin-Modified Proteome of SARS-CoV-2-Infected Host Cells Reveals Insights into Virus-Host Interaction and Pathogenesis. J. Proteome Res. 2021, 20, 2224–2239. [Google Scholar] [CrossRef] [PubMed]
- Renaudin, X.; Guervilly, J.H.; Aoufouchi, S.; Rosselli, F. Proteomic analysis reveals a FANCA-modulated neddylation pathway involved in CXCR5 membrane targeting and cell mobility. J. Cell Sci. 2014, 127, 3546–3554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bian, Y.; Song, C.; Cheng, K.; Dong, M.; Wang, F.; Huang, J.; Sun, D.; Wang, L.; Ye, M.; Zou, H. An enzyme assisted RP-RPLC approach for in-depth analysis of human liver phosphoproteome. J. Proteom. 2014, 96, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Deeb, S.J.; Cox, J.; Schmidt-Supprian, M.; Mann, M. N-linked glycosylation enrichment for in-depth cell surface proteomics of diffuse large B-cell lymphoma subtypes. Mol. Cell Proteom. 2014, 13, 240–251. [Google Scholar] [CrossRef] [Green Version]
- Bordoli, M.R.; Yum, J.; Breitkopf, S.B.; Thon, J.N.; Italiano, J.E., Jr.; Xiao, J.; Worby, C.; Wong, S.K.; Lin, G.; Edenius, M.; et al. A secreted tyrosine kinase acts in the extracellular environment. Cell 2014, 158, 1033–1044. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cabana, V.C.; Lussier, M.P. From Drosophila to Human: Biological Function of E3 Ligase Godzilla and Its Role in Disease. Cells 2022, 11, 380. https://doi.org/10.3390/cells11030380
Cabana VC, Lussier MP. From Drosophila to Human: Biological Function of E3 Ligase Godzilla and Its Role in Disease. Cells. 2022; 11(3):380. https://doi.org/10.3390/cells11030380
Chicago/Turabian StyleCabana, Valérie C., and Marc P. Lussier. 2022. "From Drosophila to Human: Biological Function of E3 Ligase Godzilla and Its Role in Disease" Cells 11, no. 3: 380. https://doi.org/10.3390/cells11030380