Bidirectional Microbiome-Gut-Brain-Axis Communication Influences Metabolic Switch-Associated Responses in the Mosquito Anopheles culicifacies
Abstract
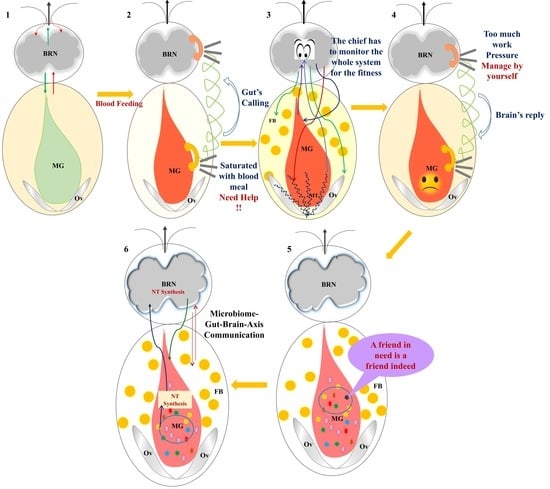
:1. Introduction
2. Material and Methods
2.1. Mosquito Rearing and Maintenance
2.2. RNA Isolation and Transcriptome Sequencing Analysis
2.3. PCR-Based Gene Expression Analysis
2.4. ROS Determination Assay of Blood-Fed Mosquitos’ Brain
2.5. Antibiotic Treatment of Mosquitoes
2.6. Decapitation Experiment
2.7. Sample Processing and MS Analysis for Neurotransmitter Quantification
2.8. dsRNA-Mediated Gene Silencing
2.9. Metagenomics Analysis & Microbiome Profiling
3. Results
3.1. Blood Meal Ingestion Enhances the Brain’s Energy Metabolism
3.2. Spatial and Temporal Modulations of Neuro-Signaling Influence Metabolic Switch-Associated Physiological Activities
3.3. Innate Physiological Status Differentially Modulates Tissue-Specific Neuromodulators/Receptors Transcripts Expression
3.4. Gut, as the ‘Second Brain’ Communicates the Nutritional Status through Neurotransmitter Synthesis
3.5. Symbiotic Gut Flora Influences Gut-Brain Axis Communication
4. Discussion
4.1. Gut-Metabolic Switch Modulates the Brain’s Energy Metabolism and Functional Engagement
4.2. Neuromodulatory Responses Establish Brain-Distant Organ Communication
4.3. Neurotransmitter Signaling and Microbiome Alteration Influences Gut-Brain-Axis Communications
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Das De, T.; Thomas, T.; Verma, S.; Singla, D.; Chauhan, C.; Srivastava, V.; Sharma, P.; Kumari, S.; Tevatiya, S.; Rani, J.; et al. A synergistic transcriptional regulation of olfactory genes drives blood-feeding associated complex behavioral responses in the mosquito anopheles culicifacies. Front. Physiol. 2018, 9, 577. [Google Scholar] [CrossRef] [PubMed]
- Potter, C.J. Stop the biting: Targeting a mosquito’s sense of smell. Cell 2014, 156, 878–881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das De, T.; Dixit, R. Neuro-Olfactory Regulation and Salivary Actions: A Coordinated Event for Successful Blood-Feeding Behavior of Mosquitoes. In Sino-Nasal and Olfactory System Disorders; InTech Open: London, UK, 2020. [Google Scholar]
- Duvall, L.B.; Ramos-Espiritu, L.; Barsoum, K.E.; Glickman, J.F.; Vosshall, L.B. Small-Molecule Agonists of Ae. aegypti Neuropeptide Y Receptor Block Mosquito Biting. Cell 2019, 176, 687–701.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beyenbach, K.; Petzel, D. Diuresis in Mosquitoes: Role of a Natriuretic Factor. Physiology 2017, 2, 171–175. [Google Scholar] [CrossRef]
- Beyenbach, K.W. A dynamic paracellular pathway serves diuresis in mosquito Malpighian tubules. Ann. N. Y. Acad. Sci. 2012, 1258, 166–176. [Google Scholar] [CrossRef] [Green Version]
- Sanders, H.R.; Evans, A.M.; Ross, L.S.; Gill, S.S. Blood meal induces global changes in midgut gene expression in the disease vector, Aedes aegypti. Insect Biochem. Mol. Biol. 2003, 33, 1105–1122. [Google Scholar] [CrossRef]
- Badisco, L.; Van Wielendaele, P.V.; Broeck, J. Vanden Eat to reproduce: A key role for the insulin signaling pathway in adult insects. Front. Physiol. 2013, 4, 202. [Google Scholar] [CrossRef] [Green Version]
- Liu, Q.; Jin, L.H. Organ-to-Organ Communication: A Drosophila Gastrointestinal Tract Perspective. Front. Cell Dev. Biol. 2017, 5, 29. [Google Scholar] [CrossRef] [Green Version]
- Duvall, L.B. Mosquito Host-Seeking Regulation: Targets for Behavioral Control. Trends Parasitol. 2019, 35, 704–714. [Google Scholar] [CrossRef]
- Droujinine, I.A.; Perrimon, N. Interorgan Communication Pathways in Physiology: Focus on Drosophila. Annu. Rev. Genet. 2016, 50, 539–570. [Google Scholar] [CrossRef] [Green Version]
- Gulia-Nuss, M.; Robertson, A.E.; Brown, M.R.; Strand, M.R. Insulin-like peptides and the target of rapamycin pathway coordinately regulate blood digestion and egg maturation in the mosquito Aedes aegypti. PLoS ONE 2011, 6, e20401. [Google Scholar] [CrossRef] [PubMed]
- Fadda, M.; Hasakiogullari, I.; Temmerman, L.; Beets, I.; Zels, S.; Schoofs, L. Regulation of Feeding and Metabolism by Neuropeptide F and Short Neuropeptide F in Invertebrates. Front. Endocrinol. 2019, 10, 64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christ, P.; Reifenrath, A.; Kahnt, J.; Hauser, F.; Hill, S.R.; Schachtner, J.; Ignell, R. Feeding-induced changes in allatostatin-A and short neuropeptide F in the antennal lobes affect odor-mediated host seeking in the yellow fever mosquito, Aedes aegypti. PLoS ONE 2017, 12, e0188243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mayer, E.A. Gut feelings: The emerging biology of gut-"brain communication. Nat. Rev. Neurosci. 2011, 12, 453–466. [Google Scholar] [CrossRef] [PubMed]
- Ridaura, V.; Belkaid, Y. Gut microbiota: The link to your second brain. Cell 2015, 161, 193–194. [Google Scholar] [CrossRef] [Green Version]
- Fülling, C.; Dinan, T.G.; Cryan, J.F. Gut Microbe to Brain Signaling: What Happens in Vagus. Neuron 2019, 101, 998–1002. [Google Scholar] [CrossRef] [Green Version]
- Forsythe, P.; Kunze, W.A. Voices from within: Gut microbes and the CNS. Cell. Mol. Life Sci. 2013, 70, 55–69. [Google Scholar] [CrossRef]
- Lampe, L.; Jentzsch, M.; Kierszniowska, S.; Levashina, E.A. Metabolic balancing by miR-276 shapes the mosquito reproductive cycle and Plasmodium falciparum development. Nat. Commun. 2019, 10, 5634. [Google Scholar] [CrossRef] [Green Version]
- Romoli, O.; Gendrin, M. The tripartite interactions between the mosquito, its microbiota and Plasmodium. Parasites Vectors 2018, 11, 200. [Google Scholar] [CrossRef] [Green Version]
- Sharma, P.; Sharma, S.; Mishra, A.K.; Thomas, T.; Das De, T.; Rohilla, S.L.; Singh, N.; Pandey, K.C.; Valecha, N.; Dixit, R. Unraveling dual feeding associated molecular complexity of salivary glands in the mosquito Anopheles culicifacies. Biol. Open 2015, 4, 1002–1015. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Natarajan, N.; Ramakrishnan, P.; Lakshmanan, V.; Palakodeti, D.; Rangiah, K. A quantitative metabolomics peek into planarian regeneration. Analyst 2015, 140, 3445–3464. [Google Scholar] [CrossRef] [PubMed]
- Ramesh, D.; Brockmann, A. Mass Spectrometric Quantification of Arousal Associated Neurochemical Changes in Single Honey Bee Brains and Brain Regions. ACS Chem. Neurosci. 2019, 10, 1950–1959. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Rani, J.; Chauhan, C.; Kumari, S.; Tevatiya, S.; Das De, T.; Savargaonkar, D.; Pandey, K.C.; Dixit, R. Altered Gut Microbiota and Immunity Defines Plasmodium vivax Survival in Anopheles stephensi. Front. Immunol. 2020, 11, 609. [Google Scholar] [CrossRef] [PubMed]
- Götz, S.; García-Gómez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talón, M.; Dopazo, J.; Conesa, A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef]
- De, T.D.; Sharma, P.; Rawal, C.; Kumari, S.; Tavetiya, S.; Yadav, J.; Hasija, Y.; Dixit, R. Sex specific molecular responses of quick-to-court protein in Indian malarial vector Anopheles culicifacies: Conflict of mating versus blood feeding behaviour. Heliyon 2017, 3, e00361. [Google Scholar] [CrossRef] [Green Version]
- Das De, T.; Hasija, Y.; Dixit, R. Transcriptional responses of attractin gene in the mosquito Anopheles culicifacies: A synergistic neuro-olfactory regulation. J. Vector Borne Dis. 2018, 55, 89–97. [Google Scholar] [CrossRef]
- Austin, S.; St-Pierre, J. PGC1 and mitochondrial metabolism—Emerging concepts and relevance in ageing and neurodegenerative disorders. J. Cell Sci. 2012, 125, 4963–4971. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.; Handschin, C.; Spiegelman, B.M. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005, 1, 361–370. [Google Scholar] [CrossRef] [Green Version]
- Liang, H.; Ward, W.F. PGC-1α: A key regulator of energy metabolism. Adv. Physiol. Educ. 2006, 30, 145–151. [Google Scholar] [CrossRef]
- Shukla, E.; Thorat, L.J.; Nath, B.B.; Gaikwad, S.M. Insect trehalase: Physiological significance and potential applications. Glycobiology 2015, 25, 357–367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mattila, J.; Hietakangas, V. Regulation of carbohydrate energy metabolism in Drosophila melanogaster. Genetics 2017, 207, 1231–1253. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Wang, X.L.; Saha, T.T.; Roy, S.; Zhao, B.; Raikhel, A.S.; Zou, Z. Temporal Coordination of Carbohydrate Metabolism during Mosquito Reproduction. PLoS Genet. 2015, 11, e1005309. [Google Scholar] [CrossRef] [PubMed]
- Sim, C.; Denlinger, D.L. A shut-down in expression of an insulin-like peptide, ILP-1, halts ovarian maturation during the overwintering diapause of the mosquito Culex pipiens. Insect Mol. Biol. 2009, 18, 325–332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kersch, C.N.; Pietrantonio, P.V. Mosquito Aedes aegypti (L.) leucokinin receptor is critical for in vivo fluid excretion post blood feeding. FEBS Lett. 2011, 585, 3507–3512. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Denlinger, D.L. Molecular structure of the prothoracicotropic hormone gene in the northern house mosquito, Culex pipiens, and its expression analysis in association with diapause and blood feeding. Insect Mol. Biol. 2011, 20, 201–213. [Google Scholar] [CrossRef] [Green Version]
- Liesch, J.; Bellani, L.L.; Vosshall, L.B. Functional and Genetic Characterization of Neuropeptide Y-Like Receptors in Aedes aegypti. PLoS Negl. Trop. Dis. 2013, 7, e2486. [Google Scholar] [CrossRef] [Green Version]
- Strand, M.R.; Brown, M.R.; Vogel, K.J. Mosquito Peptide Hormones: Diversity, Production, and Function. In Advances in Insect Physiology; Academic Press: Cambridge, MA, USA, 2016; ISBN 9780128024577. [Google Scholar]
- Piermarini, P.M.; Esquivel, C.J.; Denton, J.S. Malpighian tubules as novel targets for mosquito control. Int. J. Environ. Res. Public Health 2017, 14, 111. [Google Scholar] [CrossRef] [Green Version]
- Kinney, M.P.; Panting, N.D.; Clark, T.M. Modulation of appetite and feeding behavior of the larval mosquito Aedes aegypti by the serotonin-selective reuptake inhibitor paroxetine: Shifts between distinct feeding modes and the influence of feeding status. J. Exp. Biol. 2014, 217, 935–943. [Google Scholar] [CrossRef] [Green Version]
- Ling, L.; Raikhel, A.S. Serotonin signaling regulates insulin-like peptides for growth, reproduction, and metabolism in the disease vector Aedes aegypti. Proc. Natl. Acad. Sci. USA 2018, 115, E9822–E9831. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.; Man, Y.; Li, J.; Pei, D.; Wu, W. Olfactory ionotropic receptors in mosquito aedes albopictus (Diptera: Culicidae). J. Med. Entomol. 2017, 54, 1229–1235. [Google Scholar] [CrossRef] [PubMed]
- Bowery, N.G.; Smart, T.G. GABA and glycine as neurotransmitters: A brief history. Br. J. Pharmacol. 2006, 147 (Suppl. S1), S109–S119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hansen, I.A.; Attardo, G.M.; Rodriguez, S.D.; Drake, L.L. Four-way regulation of mosquito yolk protein precursor genes by juvenile hormone-, ecdysone-, nutrient-, and insulin-like peptide signaling pathways. Front. Physiol. 2014, 5, 103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arsic, D.; Guerin, P.M. Nutrient content of diet affects the signaling activity of the insulin/target of rapamycin/p70 S6 kinase pathway in the African malaria mosquito Anopheles gambiae. J. Insect Physiol. 2008, 54, 1226–1235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coast, G.M. Mosquito natriuretic peptide identified as a calcitonin-like diuretic hormone in Anopheles gambiae (Giles). J. Exp. Biol. 2005, 208, 3281–3291. [Google Scholar] [CrossRef] [Green Version]
- Brown, M.R.; Clark, K.D.; Gulia, M.; Zhao, Z.; Garczynski, S.F.; Crim, J.W.; Suderman, R.J.; Strand, M.R. An insulin-like peptide regulates egg maturation and metabolism in the mosquito Aedes aegypti. Proc. Natl. Acad. Sci. USA 2008, 105, 5716–5721. [Google Scholar] [CrossRef] [Green Version]
- Sharma, A.; Nuss, A.B.; Gulia-Nuss, M. Insulin-like peptide signaling in mosquitoes: The road behind and the road ahead. Front. Endocrinol. 2019, 10, 166. [Google Scholar] [CrossRef] [Green Version]
- Vogel, K.J.; Brown, M.R.; Strand, M.R. Ovary ecdysteroidogenic hormone requires a receptor tyrosine kinase to activate egg formation in the mosquito Aedes aegypti. Proc. Natl. Acad. Sci. USA 2015, 112, 5057–5062. [Google Scholar] [CrossRef] [Green Version]
- Solari, P.; Rivelli, N.; De Rose, F.; Picciau, L.; Murru, L.; Stoffolano, J.G.; Liscia, A. Opposite effects of 5-HT/AKH and octopamine on the crop contractions in adult Drosophila melanogaster: Evidence of a double brain-gut serotonergic circuitry. PLoS ONE 2017, 12, e0174172. [Google Scholar] [CrossRef] [Green Version]
- Guégan, M.; Zouache, K.; Démichel, C.; Minard, G.; Tran Van, V.; Potier, P.; Mavingui, P.; Valiente Moro, C. The mosquito holobiont: Fresh insight into mosquito-microbiota interactions. Microbiome 2018, 6, 49. [Google Scholar] [CrossRef]
- O’Mahony, S.M.; Clarke, G.; Borre, Y.E.; Dinan, T.G.; Cryan, J.F. Serotonin, tryptophan metabolism and the brain-gut-microbiome axis. Behav. Brain Res. 2015, 277, 32–48. [Google Scholar] [CrossRef] [PubMed]
- Matthews, B.J.; McBride, C.S.; DeGennaro, M.; Despo, O.; Vosshall, L.B. The neurotranscriptome of the Aedes aegypti mosquito. BMC Genom. 2016, 17, 32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plaçais, P.Y.; De Tredern, É.; Scheunemann, L.; Trannoy, S.; Goguel, V.; Han, K.A.; Isabel, G.; Preat, T. Upregulated energy metabolism in the Drosophila mushroom body is the trigger for long-term memory. Nat. Commun. 2017, 8, 15510. [Google Scholar] [CrossRef] [Green Version]
- Oriach, C.S.; Robertson, R.C.; Stanton, C.; Cryan, J.F.; Dinan, T.G. Food for thought: The role of nutrition in the microbiota-gut-brain axis. Clin. Nutr. Exp. 2016, 6, 25–38. [Google Scholar] [CrossRef] [Green Version]
- Yellen, G. Fueling thought: Management of glycolysis and oxidative phosphorylation in neuronal metabolism. J. Cell Biol. 2018, 217, 2235–2246. [Google Scholar] [CrossRef] [PubMed]
- Roh, E.; Song, D.K.; Kim, M.S. Emerging role of the brain in the homeostatic regulation of energy and glucose metabolism. Exp. Mol. Med. 2016, 48, e216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Volkenhoff, A.; Weiler, A.; Letzel, M.; Stehling, M.; Klämbt, C.; Schirmeier, S. Glial glycolysis is essential for neuronal survival in drosophila. Cell Metab. 2015, 22, 437–447. [Google Scholar] [CrossRef] [Green Version]
- Murphy, K.R.; Deshpande, S.A.; Yurgel, M.E.; Quinn, J.P.; Weissbach, J.L.; Keene, A.C.; Dawson-Scully, K.; Huber, R.; Tomchik, S.M.; Ja, W.W. Postprandial sleep mechanics in Drosophila. eLife 2016, 5, e19334. [Google Scholar] [CrossRef]
- Mittal, R.; Debs, L.H.; Patel, A.P.; Nguyen, D.; Patel, K.; O’Connor, G.; Grati, M.; Mittal, J.; Yan, D.; Eshraghi, A.A.; et al. Neurotransmitters: The Critical Modulators Regulating Gut–Brain Axis. J. Cell. Physiol. 2017, 232, 2359–2372. [Google Scholar] [CrossRef] [Green Version]
- Holzer, P.; Farzi, A. Neuropeptides and the microbiota- Gut-brain axis. In Advances in Experimental Medicine and Biology; Springer: Berlin/Heidelberg, Germany, 2014. [Google Scholar] [CrossRef] [Green Version]
- Imamura, T.; Baldwin, T.O.; Riggs, A. The amino acid sequence of the monomeric hemoglobin component from the bloodworm, Glyat liver. J. Biol. Chem. 1972, 247, 2785–2797. [Google Scholar] [CrossRef]
- Sterkel, M.; Perdomo, H.D.; Guizzo, M.G.; Barletta, A.B.F.; Nunes, R.D.; Dias, F.A.; Sorgine, M.H.F.; Oliveira, P.L. Tyrosine Detoxification Is an Essential Trait in the Life History of Blood-Feeding Arthropods. Curr. Biol. 2016, 26, 2188–2193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- French, A.S.; Simcock, K.L.; Rolke, D.; Gartside, S.E.; Blenau, W.; Wright, G.A. The role of serotonin in feeding and gut contractions in the honeybee. J. Insect Physiol. 2014, 61, 8–15. [Google Scholar] [CrossRef] [Green Version]
- Kamhi, J.F.; Arganda, S.; Moreau, C.S.; Traniello, J.F.A. Origins of Aminergic Regulation of Behavior in Complex Insect Social Systems. Front. Syst. Neurosci. 2017, 11, 74. [Google Scholar] [CrossRef] [Green Version]
- Friedman, D.A.; Pilko, A.; Skowronska-Krawczyk, D.; Krasinska, K.; Parker, J.W.; Hirsh, J.; Gordon, D.M. The Role of Dopamine in the Collective Regulation of Foraging in Harvester Ants. iScience 2018, 8, 283–294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vinauger, C.; Lahondère, C.; Wolff, G.H.; Locke, L.T.; Liaw, J.E.; Parrish, J.Z.; Akbari, O.S.; Dickinson, M.H.; Riffell, J.A. Modulation of Host Learning in Aedes aegypti Mosquitoes. Curr. Biol. 2018, 28, 333–344.e8. [Google Scholar] [CrossRef] [Green Version]
- Vinauger, C. Vector cognition and neurobiology. Curr. Opin. Insect Sci. 2019, 34, 68–72. [Google Scholar] [CrossRef] [PubMed]
- Ngai, M.; Shoue, D.A.; Loh, Z.; McDowell, M.A. The pharmacological and functional characterization of the serotonergic system in Anopheles gambiae and Aedes aegypti: Influences on flight and blood-feeding behavior. Sci. Rep. 2019, 9, 4421. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.K.; Kim, Y.S.; Lee, H.M.; Jin, H.S.; Neupane, C.; Kim, S.; Lee, S.H.; Min, J.J.; Sasai, M.; Jeong, J.H.; et al. GABAergic signaling linked to autophagy enhances host protection against intracellular bacterial infections. Nat. Commun. 2018, 9, 4184. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Zhang, R.; Zhang, B.; Zhao, T.; Wang, P.; Liang, G.; Cheng, G. Blood meal acquisition enhances arbovirus replication in mosquitoes through activation of the GABAergic system. Nat. Commun. 2017, 8, 1262. [Google Scholar] [CrossRef] [Green Version]
- Sarkar, A.; Lehto, S.M.; Harty, S.; Dinan, T.G.; Cryan, J.F.; Burnet, P.W.J. Psychobiotics and the Manipulation of Bacteria–Gut–Brain Signals. Trends Neurosci. 2016, 39, 763–781. [Google Scholar] [CrossRef] [Green Version]
- Montiel-Castro, A.J.; González-Cervantes, R.M.; Bravo-Ruiseco, G.; Pacheco-López, G. The microbiota-gut-brain axis: Neurobehavioral correlates, health and sociality. Front. Integr. Neurosci. 2013, 7, 70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parker, A.; Fonseca, S.; Carding, S.R. Gut microbes and metabolites as modulators of blood-brain barrier integrity and brain health. Gut Microbes 2020, 11, 135–157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van de Wouw, M.; Schellekens, H.; Dinan, T.G.; Cryan, J.F. Microbiota-Gut-Brain Axis: Modulator of Host Metabolism and Appetite. J. Nutr. 2017, 147, 727–745. [Google Scholar] [CrossRef] [Green Version]
- Kawase, T.; Nagasawa, M.; Ikeda, H.; Yasuo, S.; Koga, Y.; Furuse, M. Gut microbiota of mice putatively modifies amino acid metabolism in the host brain. Br. J. Nutr. 2017, 117, 775–783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Westfall, S.; Lomis, N.; Prakash, S. Longevity extension in Drosophila through gut-brain communication. Sci. Rep. 2018, 8, 8362. [Google Scholar] [CrossRef] [PubMed]
- Akami, M.; Andongma, A.A.; Zhengzhong, C.; Nan, J.; Khaeso, K.; Jurkevitch, E.; Niu, C.Y.; Yuval, B. Intestinal bacteria modulate the foraging behavior of the oriental fruit fly Bactrocera dorsalis (Diptera: Tephritidae). PLoS ONE 2019, 14, e0210109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gareau, M.G. Microbiota-gut-brain axis and cognitive function. In Advances in Experimental Medicine and Biology; Springer: Berlin/Heidelberg, Germany, 2014. [Google Scholar] [CrossRef]
- Blumberg, B.J.; Short, S.M.; Dimopoulos, G. Employing the Mosquito Microflora for Disease Control. In Genetic Control of Malaria and Dengue; Academic Press: Cambridge, MA, USA, 2015; ISBN 9780128004050. [Google Scholar]
- Kaur, H.; Bose, C.; Mande, S.S. Tryptophan Metabolism by Gut Microbiome and Gut-Brain-Axis: An in silico Analysis. Front. Neurosci. 2019, 13, 1365. [Google Scholar] [CrossRef]
- Jenkins, T.A.; Nguyen, J.C.D.; Polglaze, K.E.; Bertrand, P.P. Influence of tryptophan and serotonin on mood and cognition with a possible role of the gut-brain axis. Nutrients 2016, 8, 56. [Google Scholar] [CrossRef]
- Taylor, P.; Brown, J.H. Basic Neurochemistry: Molecular, Cellular and Medical Aspects. In Basic Neurochemistry: Molecular, Cellular and Medical Aspects, 6th ed.; Academic Press: Cambridge, MA, USA, 1943. [Google Scholar]
- Chen, T.T.; Strahlendorf, P.W.; Wyatt, G.R. Vitellin and vitellogenin from locusts (Locusta migratoria). J. Biol. Chem. 1978, 253, 5325–5331. [Google Scholar] [CrossRef]
- Gaio, A.D.O.; Gusmão, D.S.; Santos, A.V.; Berbert-Molina, M.A.; Pimenta, P.F.P.; Lemos, F.J.A. Contribution of midgut bacteria to blood digestion and egg production in aedes aegypti (diptera: Culicidae) (L.). Parasites Vectors 2011, 4, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Sudo, N.; Chida, Y.; Aiba, Y.; Sonoda, J.; Oyama, N.; Yu, X.N.; Kubo, C.; Koga, Y. Postnatal microbial colonization programs the hypothalamic-pituitary-adrenal system for stress response in mice. J. Physiol. 2004, 558, 263–275. [Google Scholar] [CrossRef] [PubMed]
- Mazzoli, R.; Pessione, E. The neuro-endocrinological role of microbial glutamate and GABA signaling. Front. Microbiol. 2016, 7, 1934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De, T.D.; Sharma, P.; Thomas, T.; Singla, D.; Tevatiya, S.; Kumari, S.; Chauhan, C.; Rani, J.; Srivastava, V.; Kaur, R.; et al. Interorgan molecular communication strategies of “Local” and “Systemic” innate immune responses in mosquito Anopheles stephensi. Front. Immunol. 2018, 9, 148. [Google Scholar] [CrossRef] [Green Version]
- Chen, A.; Singh, C.; Oikonomou, G.; Prober, D.A. Genetic Analysis of Histamine Signaling in Larval Zebrafish Sleep. eNeuro 2017, 4, ENEURO.0286-16.2017. [Google Scholar] [CrossRef] [PubMed]
- Bushey, D.; Tononi, G.; Cirelli, C. Sleep- and wake-dependent changes in neuronal activity and reactivity demonstrated in fly neurons using in vivo calcium imaging. Proc. Natl. Acad. Sci. USA 2015, 112, 4785–4790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cava, F.; Lam, H.; De Pedro, M.A.; Waldor, M.K. Emerging knowledge of regulatory roles of d-amino acids in bacteria. Cell. Mol. Life Sci. 2011, 68, 817–831. [Google Scholar] [CrossRef] [Green Version]
- Aliashkevich, A.; Alvarez, L.; Cava, F. New insights into the mechanisms and biological roles of D-amino acids in complex eco-systems. Front. Microbiol. 2018, 9, 683. [Google Scholar] [CrossRef] [Green Version]






| Sl. No. | Gene Name | Synthesized from | Target Tissue | Possible Function | Target Tissue for Expression Study |
|---|---|---|---|---|---|
| 1. | ILP1 | MNSC of brain | Multiple tissues | Halt ovarian maturation [35] | Brain, midgut |
| 2. | ILP3 | MNSC of brain | Midgut, Ovary, Fat Body, Hemocyte | Nutrient storage by FB, regulation of digestive enzymes by MG, Ecdysteroid production from ovaries, the immune response by HC [8,36] | Brain, midgut |
| 3. | Leucokinin | Abdominal ganglia | Gut, Malpighian tubule | Regulation of fluid secretion, ionic balance [36] | Brain |
| 4. | PTTH—Prothoracicotropic Hormone | Brain | Not Known | Diapause and blood-feeding [37] | Brain |
| 5. | Neuropeptide Y Receptor—NRY | NSC of brain | Brain | Host-seeking inhibition [4,38] | Brain |
| 6. | Leucokinin Receptor | Multiple tissues | Multiple tissues | Regulation of fluid secretion, ionic balance [39] | Brain, midgut |
| 7. | Diuretic hormone 44 (DH44) | Gut endocrine cells | Malpighian tubule | Regulation of diuresis [40] | Brain, midgut |
| 8. | OEH—Ovary Ecdysteroidogenic Hormone | MNSC and ventricular ganglia of the brain | Ovary | Induces ecdysone production from the ovary after blood feeding [39] | Brain |
| 9. | ARMAA—Aromatic-L-amino-acid decarboxylase | Multiple tissues | Multiple tissues | Synthesis of serotonin neurotransmitter and regulation of multiple physiological processes | Brain |
| 10. | DH44R1 | Malpighian tubule | Malpighian tubule | Regulation of Diuresis [39,40] | Midgut and Malpighian tubule |
| 11. | CCHamide Receptor 2 | CCHamide2 synthesized from gut endocrine cells | Multiple tissues | Nutrient dependent regulation of ILPs from brain [39] | Midgut |
| 12. | 5HTR—Serotonin Receptor | Multiple tissues | Multiple tissues | Multiple behavioral and physiological processes [41,42] | Brain, Midgut |
| 13. | Glutamate R—Glutamate Receptor | Multiple tissues | Multiple tissues | Olfactory ionotropic glutamate receptor in odorant recognition (Identified from AC brain transcriptome data) [43] | Brain, Midgut |
| 14. | Glycine R—Glycine Receptor | Multiple tissues | Multiple tissues | Inhibit neurotransmission (Identified from AC brain transcriptome data) [44] | Brain, Midgut |
| 15. | Akt Kinase—Protein kinase B | Multiple | Multiple | Activation of TOR pathway [8] | Ovary |
| 16. | CYP31A41-20E hydroxylase (20E synthesizing enzyme) | Ovary | Fat body and ovary | Ovary and oocyte development [45] | Ovary |
| 17. | STPK—Serine threonine-protein kinase | Multiple | Multiple | Multiple physiological processes [46] | Brain, Ovary |
| 18. | PI4-Kinase | Multiple | Multiple | Multiple physiological processes (Identified from AC brain transcriptome data) | Brain, Ovary |
| 19. | Calcitonin Receptor | Malphigian tubule | Malphigian tubule | Regulation of diuresis [40,47] | Malphigian tubule |
| 20. | KDNaCa Exchanger | Malpighian tubule | Malpighian tubule | Regulate fluid secretion and diuresis [40] | Malpighian tubule |
| 21. | V-Type ATPase | Malpighian tubule | Malpighian tubule | Regulate membrane potential and diuresis [40] | Malpighian tubule |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Das De, T.; Sharma, P.; Tevatiya, S.; Chauhan, C.; Kumari, S.; Yadav, P.; Singla, D.; Srivastava, V.; Rani, J.; Hasija, Y.; et al. Bidirectional Microbiome-Gut-Brain-Axis Communication Influences Metabolic Switch-Associated Responses in the Mosquito Anopheles culicifacies. Cells 2022, 11, 1798. https://doi.org/10.3390/cells11111798
Das De T, Sharma P, Tevatiya S, Chauhan C, Kumari S, Yadav P, Singla D, Srivastava V, Rani J, Hasija Y, et al. Bidirectional Microbiome-Gut-Brain-Axis Communication Influences Metabolic Switch-Associated Responses in the Mosquito Anopheles culicifacies. Cells. 2022; 11(11):1798. https://doi.org/10.3390/cells11111798
Chicago/Turabian StyleDas De, Tanwee, Punita Sharma, Sanjay Tevatiya, Charu Chauhan, Seena Kumari, Pooja Yadav, Deepak Singla, Vartika Srivastava, Jyoti Rani, Yasha Hasija, and et al. 2022. "Bidirectional Microbiome-Gut-Brain-Axis Communication Influences Metabolic Switch-Associated Responses in the Mosquito Anopheles culicifacies" Cells 11, no. 11: 1798. https://doi.org/10.3390/cells11111798