Candidate Genes Involved in Tolerance to Fenoxaprop-P-Ethyl in Rice Induced by Isoxadifen-Ethyl Hydrolysate
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. Seedling Treatment and Determination of Physiological Indices
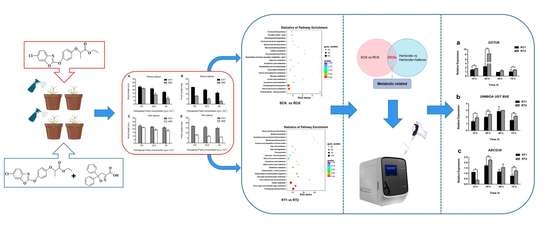
2.3. Transcriptome Profiling of Rice Seedlings in Different Treatments
2.3.1. Transcriptome Sequencing
2.3.2. Differentially Expressed Gene (DEG) Analysis and Identification of FE Metabolism-Related Genes
2.3.3. Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR) Validation
2.4. Statistical Analysis
3. Results
3.1. Bioactivity of the Whole Plant Assay
3.2. Transcriptome Sequencing and Analysis
3.3. Functional Annotation of Differentially Expressed Genes (DEGs)
3.4. qRT-PCR
3.5. Identification and Validation of Potential Fenoxaprop-P-ethyl Metabolic Genes
3.6. The Expression Patterns of DEGs after Treated with Fenoxaprop-P-ethyl
3.7. Revalidation of Potential Metabolic Genes by Safener
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sardrood, B.P.; Goltapeh, E.M. Weeds, herbicides and plant disease management. Biocontrol 2018, 31, 41–178. [Google Scholar]
- Obiri, B.D.; Obeng, E.A.; Oduro, K.A.; Apetorgbor, M.M.; Peprah, T.; Duah-Gyamfi, A.; Mensah, J.K. Farmers’ perceptions of herbicide usage in forest landscape restoration programs in Ghana. Sci. Afr. 2021, 11, e00672. [Google Scholar] [CrossRef]
- Jajoria, D.K.; Narolia, G.P.; Massey, J.X.; Singh, H. Bio-efficacy of fenoxaprop-p-ethyl 9 EC for grassy weed control in groundnut (Arachis hypogaea). Int. J. Plant Sci. 2014, 9, 266–270. [Google Scholar]
- Wu, C.X.; Wang, Q.; Zhao, X.P.; L.P, C. Safety and weed activity of fenoxaprop-P-ethyl on different rice varieties. In Public Plant Protection and Green Prevention and Control; China Agricultural Science and Technology Press: Hebi, China, 2010; pp. 396–401. (In Chinese) [Google Scholar]
- Jang, S.; Marjanovic, J.; Gornicki, P. Resistance to herbicides caused by single amino acid mutations in acetyl-CoA carboxylase in resistant populations of grassy weeds. New Phytol. 2013, 197, 1110–1116. [Google Scholar] [CrossRef] [PubMed]
- Cha, T.S.; Najihah, M.G.; Sahid, I.B.; Chuah, T.S. Molecular basis for resistance to ACCase-inhibiting fluazifop in Eleusine indica from Malaysia. Pestic. Biochem. Physiol. 2014, 111, 7–13. [Google Scholar] [CrossRef]
- Délye, C.; Jasieniuk, M.; Le Corre, V. Deciphering the evolution of herbicide resistance in weeds. Trends Genet. 2013, 29, 649–658. [Google Scholar] [CrossRef]
- Kaundun, S.S. Resistance to acetyl-CoA carboxylase-inhibiting herbicides. Pest Manag. Sci. 2014, 70, 1405–1417. [Google Scholar] [CrossRef]
- Zhao, N.; Yang, J.; Jiang, M.; Liao, M.; Cao, H. Identification of essential genes involved in metabolism-based resistance mechanism to fenoxaprop-P-ethyl in Polypogon fugax. Pest Manag. Sci. 2022, 78, 1164–1175. [Google Scholar] [CrossRef]
- Cutti, L.; Rigon, C.A.G.; Girelli, N.; Angonese, P.S.; Ulguim, A.D.R.; Merotto, A. The safener isoxadifen-ethyl confers fenoxaprop-p-ethyl resistance on a biotype of Echinochloa crus-galli. Pest Manag. Sci. 2022, 78, 2287–2298. [Google Scholar] [CrossRef]
- Shen, C.C.; Tang, W.W.; Zeng, D.Q.; Xu, H.L.; Su, W.C.; Wu, R.H. Isoxadifen-ethyl derivatives protect rice from fenoxaprop-P-ethyl–associated injury during the control of weedy rice. Weed Sci. 2017, 65, 579–587. [Google Scholar] [CrossRef]
- Kraehmer, H.; Laber, B.; Rosinger, C.; Schulz, A. Herbicides as weed control agents: State of the art: I. Weed control research and safener technology: The path to modern agriculture. Plant Physiol. 2014, 166, 1119–1131. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.L.; Xu, H.L.; Su, W.C.; Xue, F.; An, S.H.; Wu, R.H.; Lu, C.T. The expression of detoxification genes in two maize cultivars by interaction of isoxadifen-ethyl and nicosulfuron. Plant Physiol. Biochem. 2018, 129, 101–108. [Google Scholar] [CrossRef]
- Baghestani, M.; Ali, M.; Sabeti, P. Investigating Efficacy of MaisTer 6.1% OD (Foramsulfuron + Iodosulfuron + Isoxadifen-Ethyl) on Weeds in Corn; Iranian Research Institute of Plant Protection: Tehran, Iran, 2012. (in Kermanshah) [Google Scholar]
- Holt, D.C.; Lay, V.J.; Clarke, E.D.; Dinsmore, A.; Jepson, I.; Bright, S.W.; Greenland, A.J. Characterization of the safener-induced glutathione S-transferase isoform II from maize. Planta 1995, 196, 295–302. [Google Scholar] [CrossRef]
- McGonigle, B.; Keeler, S.J.; Lau, S.M.C.; Koeppe, M.K.; O’Keefe, D.P. A genomics approach to the comprehensive analysis of the glutathione S-transferase gene familyin soybean and maize. Plant Physiol. 2000, 124, 1105–1120. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Xu, F.; Lambert, K.N.; Riechers, D.E. Safeners coordinately induce the expression of multiple proteins and MRP transcripts involved in herbicide metabolism and detoxification in Triticum tauschii seedling tissues. Proteomics 2007, 7, 1261–1278. [Google Scholar] [CrossRef]
- Riechers, D.E.; Klaus, K.; Zhang, Q. Detoxification without intoxication: Herbicide safeners activate plant defense gene expression. Plant Physiol. 2010, 153, 3–13. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.N.; Li, W.Q.; Sun, L.L.; Xu, H.L.; Su, W.C.; Xue, F.; Wu, R.H.; Lu, C.T. Transcriptome analysis and the identification of genes involved in the metabolic pathways of fenoxaprop-P-ethyl in rice treated with isoxadifen-ethyl hydrolysate. Pestic. Biochem. Physiol. 2022, 183, 105057. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR. Methods 2002, 25, 402–408. [Google Scholar] [CrossRef]
- Sreedevi, B.; Krishnamurthy, P.; Singh, S.P. Effect of Herbicides on Weed Control and Nitrogen Dynamics of Direct Sown Rice and Associated Weeds; Iranian Research Institute of Plant Protection: Tehran, Iran, 2007. [Google Scholar]
- Rajkhowa, D.J.; Deka, N.C.; Borah, N.; Barua, I.C. Effect of herbicides with or without paddy weeder on weeds in transplanted summer rice (Oryza sativa). Indian J. Agron. 2007, 52, 107–110. [Google Scholar]
- Baek, Y.S.; Goodrich, L.V.; Brown, P.J.; James, B.T.; Moose, S.P.; Lambert, K.N.; Riechers, D.E. Transcriptome profiling and genome-wide association studies reveal GSTs and other defense genes involved in multiple signaling pathways induced by herbicide safener in grain sorghum. Front. Plant Sci. 2019, 10, 192. [Google Scholar] [CrossRef]
- Hu, L.F.; Yao, Y.; Cai, R.; Pan, L.; Bai, L.Y. Effects of fenclorim on rice physiology, gene transcription and pretilachlor detoxification ability. BMC Plant Biol. 2020, 20, 100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nandula, V.K.; Riechers, D.E.; Ferhatoglu, Y.; Barrett, M.; Duke, S.O.; Dayan, F.E.; Goldberg-Cavalleri, A.; Tétard-Jones, C.; Wortley, D.J.; Onkokesung, N. Herbicide metabolism: Crop selectivity, bioactivation, weed resistance, and regulation. Weed Sci. 2019, 67, 149–175. [Google Scholar] [CrossRef] [Green Version]
- Behringer, C.; Bartsch, K.; Schaller, A. Safeners recruit multiple signalling pathways for the orchestrated induction of the cellular xenobiotic detoxification machinery in Arabidopsis. Plant Cell Environ. 2011, 34, 1970–1985. [Google Scholar] [CrossRef] [PubMed]
- Dimaano, N.G.; Iwakami, S. Cytochrome P450-mediated herbicide metabolism in plants: Current understanding and prospects. Pest Manag. Sci. 2020, 77, 22–32. [Google Scholar] [CrossRef]
- Deng, F.; Hatzios, K.K. Characterization of cytochrome p450-mediated bensulfuron-methyl O-demethylation in rice. Pestic. Biochem. Physiol. 2002, 74, 102–115. [Google Scholar] [CrossRef]
- Hirose, S.; Kawahigashi, H.; Tagiri, A.; Imaishi, H.; Ohkawa, H.; Ohkawa, Y. Tissue-specific expression of rice CYP72A21 induced by auxins and herbicides. Plant Biotechnol. J. 2007, 1, 27–36. [Google Scholar] [CrossRef]
- Bai, S.; Zhao, Y.; Zhou, Y.; Wang, M.; Li, Y.; Luo, X.; Li, L. Identification and expression of main genes involved in non-target site resistance mechanisms to fenoxaprop-p-ethyl in Beckmannia syzigachne. Pest Manag. Sci. 2020, 76, 2619–2626. [Google Scholar] [CrossRef]
- Cummins, I.; Dixon, D.P.; Freitag-Pohl, S.; Skipsey, M.; Edwards, R. Multiple roles for plant glutathione transferases in xenobiotic detoxification. Drug Metab. Rev. 2011, 43, 266–280. [Google Scholar] [CrossRef]
- Dixon, D.P.; McEwen, A.G.; Lapthorn, A.J.; Edwards, R. Forced evolution of a herbicide detoxifying glutathione transferase. J. Biol. Chem. 2003, 278, 23930–23935. [Google Scholar] [CrossRef] [Green Version]
- Zou, Y.; Cao, S.; Zhao, B.; Sun, Z.; Liu, L.; Ji, M. Increase in glutathione S-transferase activity and antioxidant damage ability drive resistance to bensulfuron-methyl in Sagittaria trifolia. Plant Physiol. Biochem. 2022, 190, 240–247. [Google Scholar] [CrossRef]
- Brazier-Hicks, M.; Knight, M.K.; Sellars, D.J. Steel and Patrick, Testing a chemical series inspired by plant stress oxylipin signalling agents for herbicide safening activity. Pest Manag. Sci. 2018, 74, 828–836. [Google Scholar] [CrossRef]
- Pan, L.; Gao, H.; Xia, W.; Zhang, T.; Dong, L. Establishing a herbicide-metabolizing enzyme library in Beckmannia syzigachne to identify genes associated with metabolic resistance. J. Exp. Bot. 2016, 67, 1745–1757. [Google Scholar] [CrossRef]
- Burgos, H.N. Metabolism-based herbicide resistance: Regulation by safeners. Weed Sci. 2004, 52, 454–467. [Google Scholar]
- Huang, X.X.; Zhao, S.M.; Zhang, Y.Y.; Li, Y.J.; Shen, H.N.; Li, X.; Hou, B.K. A novel UDP-glycosyltransferase 91C1 confers specific herbicide resistance through detoxification reaction in Arabidopsis. Plant Physiol. Biochem. 2021, 159, 226–233. [Google Scholar] [CrossRef]
- Brazier, M.; Cole, D.J.; Edwards, R. O-Glucosyltransferase activities toward phenolic natural products and xenobiotics in wheat and herbicide-resistant and herbicide-susceptible black-grass (Alopecurus myosuroides). Phytochemistry 2002, 59, 149–156. [Google Scholar] [CrossRef]
- Yuan, J.S.; Tranel, P.J.; Stewart, C. Non-target-site herbicide resistance: A family business. Trends Plant Sci. 2007, 12, 6–13. [Google Scholar] [CrossRef]
- Park, Y.; Moon, Y.; Ryoo, J.; Kim, N.; Cho, H.; Ahn, J. Identification of the minimal region in lipase ABC transporter recognition domain of Pseudomonas fluorescens for secretion and fluorescence of green fluorescent protein. Microb. Cell Fact. 2012, 11, 60. [Google Scholar] [CrossRef]






| Gene ID | Accession ID | Description | GO Annotation |
|---|---|---|---|
| Os07g0635500 | XM_015789232 | CYP709B2 | GO:0005506; GO:0016021; GO:0016709; GO:0020037; GO:0044550; GO:0055114 |
| Os09g0530300 | XM_015795881 | CYP71A1 | |
| Os10g0525000 | XM_015759368 | CYP704C1 | GO:0004497; GO:0005506; GO:0009536; GO:0016021; GO:0016705; GO:0020037; GO:0022900 |
| Os01g0628700 | XM_015765608 | CYP72A11 | GO:0004497; GO:0005506; GO:0016021; GO:0016705; GO:0020037; GO:0055114 |
| Os11g0635500 | XM_015760069 | CYP71D10 | |
| Os01g0627500 | XM_015760381 | CYP72A15 | GO:0004497; GO:0005506; GO:0016021; GO:0016705; GO:0020037; GO:0022900 |
| Os08g0508000 | XM_015792583 | CYP76M5 | GO:0005506; GO:0006952; GO:0016021; GO:0016709; GO:0020037; GO:0022900; GO:0051502 |
| Os08g0465700 | XM_015794639 | CYP76M5 | GO:0005506; GO:0016021; GO:0016709; GO:0020037; GO:0022900; GO:0044550 |
| Os03g0134500 | XM_015776235 | CYP78A6 | GO:0005506; GO:0016021; GO:0016709; GO:0020037; GO:0044550; GO:0055114 |
| Os10g0527400 | XM_015758264 | GSTU6 | GO:0004364; GO:0005739; GO:0006749; GO:0009407 |
| Os10g0530900 | XM_015758870.2 | GSTU6 | GO:0004364; GO:0005737; GO:0006749; GO:0009407 |
| Os11g0441500 | XM_015762250 | DIMBOA UGT BX8 | GO:0005488; GO:0005739; GO:0008194; GO:0009813; GO:0016758; GO:0052696 |
| Os03g0757000 | XM_015775386 | UGT83A1 | GO:0009813; GO:0043231; GO:0052696; GO:0080043; GO:0080044 |
| Os04g0206700 | XM_026024995.1 | UGT79 | |
| Os06g0220500 | XM_015788488 | UGT85A2 | |
| Os01g0695800 | XM_015766599 | ABCB11 | GO:0005524; GO:0016021; GO:0042626; GO:0055085 |
| Os11g0155600 | XM_026021048 | ABCC10 | GO:0005524; GO:0016021; GO:0042626; GO:0055085 |
| Os09g0472100 | XM_015757091 | ABCG11 | GO:0005524; GO:0005886; GO:0016021; GO:0042626; GO:0055085 |
| Os02g0208300 | XM_015770762 | ABCG39 | GO:0005524; GO:0005886; GO:0009699; GO:0016021; GO:0042349; GO:0042626; GO:0050790; GO:0055085 |
| Os01g0696600 | XM_015761603.2 | ABCB4 | GO:0005524; GO:0016021; GO:0042626; GO:0055085 |
| Os01g0696701 | XM_015761603.2 | ABCB4 | |
| Os02g0190000 | XM_015769540 | MRP protein | |
| Os02g0190300 | XM_015769530 | MRP protein | |
| Os02g0634700 | XM_015769887 | SCP 34 | GO:0004185; GO:0005773; GO:0009505; GO:0051603 |
| Os07g0479300 | NM_001403059 | Serine carboxypeptidase | GO:0004185; GO:0005773; GO:0005777; GO:0005789; GO:0005829; GO:0051603 |
| Os12g0257000 | XM_015762695 | SCP 1 | GO:0004185; GO:0005576; GO:0005773; GO:0005777; GO:0016747; GO:0019748; GO:0051603 |
| Os07g0656900 | XM_015791377 | SCP II-3 | GO:0004185; GO:0016021; GO:0051603 |
| Os01g0332800 | XM_015766668 | SCP26 | GO:0004185; GO:0005773; GO:0051603 |
| Os01g0833500 | XM_015795428 | SCP27 | |
| Os06g0186400 | XM_015787453 | SCP II-2 | |
| Os08g0191700 | NM_001403516 | Lactoylglutathione lyase-like | GO:0004462; GO:0046872 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Y.; Li, W.; Sun, L.; Wu, R.; Xu, H.; Su, W.; Lu, C. Candidate Genes Involved in Tolerance to Fenoxaprop-P-Ethyl in Rice Induced by Isoxadifen-Ethyl Hydrolysate. Agronomy 2023, 13, 225. https://doi.org/10.3390/agronomy13010225
Zhao Y, Li W, Sun L, Wu R, Xu H, Su W, Lu C. Candidate Genes Involved in Tolerance to Fenoxaprop-P-Ethyl in Rice Induced by Isoxadifen-Ethyl Hydrolysate. Agronomy. 2023; 13(1):225. https://doi.org/10.3390/agronomy13010225
Chicago/Turabian StyleZhao, Yaning, Wenqing Li, Lanlan Sun, Renhai Wu, Hongle Xu, Wangcang Su, and Chuantao Lu. 2023. "Candidate Genes Involved in Tolerance to Fenoxaprop-P-Ethyl in Rice Induced by Isoxadifen-Ethyl Hydrolysate" Agronomy 13, no. 1: 225. https://doi.org/10.3390/agronomy13010225