Proteomic Analysis of the Predatory Venom of Conus striatus Reveals Novel and Population-Specific κA-Conotoxin SIVC
Abstract
:1. Introduction
2. Results
2.1. RP-HPLC of Predatory C. striatus Venoms
2.2. Mass Spectrometry Analysis of Venom Fractions
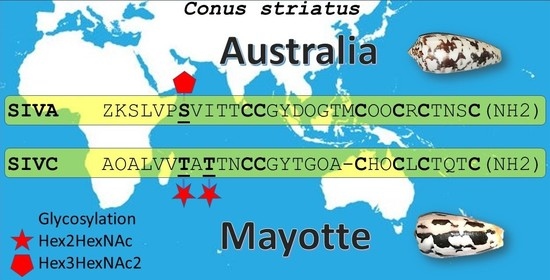
2.3. Peptide Glycosylation Investigation
2.4. De Novo Peptide Sequencing
2.5. Glycosylation Site Determination
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Materials
5.2. Venom Extraction and HPLC Fractionation
5.3. Sample Preparation
5.4. Digestion
5.5. Mass Spectrometry
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dutertre, S.; Jin, A.-H.; Vetter, I.; Hamilton, B.; Sunagar, K.; Lavergne, V.; Dutertre, V.; Fry, B.G.; Antunes, A.; Venter, D.J.; et al. Evolution of Separate Predation- and Defence-Evoked Venoms in Carnivorous Cone Snails. Nat. Commun. 2014, 5, 3521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, A.-H.; Muttenthaler, M.; Dutertre, S.; Himaya, S.W.A.; Kaas, Q.; Craik, D.J.; Lewis, R.J.; Alewood, P.F. Conotoxins: Chemistry and Biology. Chem. Rev. 2019, 119, 11510–11549. [Google Scholar] [CrossRef]
- Prashanth, J.R.; Brust, A.; Jin, A.-H.; Alewood, P.F.; Dutertre, S.; Lewis, R.J. Cone Snail Venomics: From Novel Biology to Novel Therapeutics. Futur. Med. Chem. 2014, 6, 1659–1675. [Google Scholar] [CrossRef] [PubMed]
- Lewis, R.J.; Dutertre, S.; Vetter, I.; Christie, M.J. Conus Venom Peptide Pharmacology. Pharmacol. Rev. 2012, 64, 259–298. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.; Peng, C.; Yang, J.; Yi, Y.; Zhang, J.; Shi, Q. Cone Snails: A Big Store of Conotoxins for Novel Drug Discovery. Toxins 2017, 9, 397. [Google Scholar] [CrossRef] [Green Version]
- Kaas, Q.; Yu, R.; Jin, A.-H.; Dutertre, S.; Craik, D.J. ConoServer: Updated Content, Knowledge, and Discovery Tools in the Conopeptide Database. Nucleic Acids Res. 2012, 40, D325–D330. [Google Scholar] [CrossRef]
- Terlau, H.; Shon, K.-J.; Grilley, M.; Stocker, M.; Stühmer, W.; Olivera, B.M. Strategy for Rapid Immobilization of Prey by a Fish-Hunting Marine Snail. Nature 1996, 381, 148–151. [Google Scholar] [CrossRef]
- Craig, A.G.; Zafaralla, G.; Cruz, L.J.; Santos, A.D.; Hillyard, D.R.; Dykert, J.; Rivier, J.E.; Gray, W.R.; Imperial, J.; DelaCruz, R.G.; et al. An O-Glycosylated Neuroexcitatory Conus Peptide. Biochemistry 1998, 37, 16019–16025. [Google Scholar] [CrossRef]
- Kelley, W.P.; Schulz, J.R.; Jakubowski, J.A.; Gilly, W.F.; Sweedler, J.V. Two Toxins from Conus Striatus That Individually Induce Tetanic Paralysis. Biochemistry 2006, 45, 14212–14222. [Google Scholar] [CrossRef] [Green Version]
- Jakubowski, J.A.; Kelley, W.P.; Sweedler, J.V. Screening for Post-Translational Modifications in Conotoxins Using Liquid Chromatography/Mass Spectrometry: An Important Component of Conotoxin Discovery. Toxicon 2006, 47, 688–699. [Google Scholar] [CrossRef]
- Medzihradszky, K.F.; Chalkley, R.J. Lessons in de Novo Peptide Sequencing by Tandem Mass Spectrometry. Mass Spectrom. Rev. 2015, 34, 43–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mikesh, L.M.; Ueberheide, B.; Chi, A.; Coon, J.J.; Syka, J.E.P.; Shabanowitz, J.; Hunt, D.F. The Utility of ETD Mass Spectrometry in Proteomic Analysis. Biochim. Biophys. Acta-Proteins Proteomics 2006, 1764, 1811–1822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerwig, G.; Hocking, H.; Stöcklin, R.; Kamerling, J.; Boelens, R. Glycosylation of Conotoxins. Mar. Drugs 2013, 11, 623–642. [Google Scholar] [CrossRef] [PubMed]
- Wiesner, J.; Premsler, T.; Sickmann, A. Application of Electron Transfer Dissociation (ETD) for the Analysis of Posttranslational Modifications. Proteomics 2008, 8, 4466–4483. [Google Scholar] [CrossRef]
- Syka, J.E.P.; Coon, J.J.; Schroeder, M.J.; Shabanowitz, J.; Hunt, D.F. Peptide and Protein Sequence Analysis by Electron Transfer Dissociation Mass Spectrometry. Proc. Natl. Acad. Sci. USA 2004, 101, 9528–9533. [Google Scholar] [CrossRef] [Green Version]
- Penn, S.G.; Cancilla, M.T.; Lebrilla, C.B. Collision-Induced Dissociation of Branched Oligosaccharide Ions with Analysis and Calculation of Relative Dissociation Thresholds. Anal. Chem. 1996, 68, 2331–2339. [Google Scholar] [CrossRef]
- Paizs, B.; Suhai, S. Fragmentation Pathways of Protonated Peptides. Mass Spectrom. Rev. 2005, 24, 508–548. [Google Scholar] [CrossRef]
- Santos, A.D.; McIntosh, J.M.; Hillyard, D.R.; Cruz, L.J.; Olivera, B.M. The A-Superfamily of Conotoxins. J. Biol. Chem. 2004, 279, 17596–17606. [Google Scholar] [CrossRef] [Green Version]
- Puillandre, N.; Watkins, M.; Olivera, B.M. Evolution of Conus Peptide Genes: Duplication and Positive Selection in the A-Superfamily. J. Mol. Evol. 2010, 70, 190–202. [Google Scholar] [CrossRef] [Green Version]
- Tsai, P.L.; Chen, S.-F.; Huang, S.Y. Mass Spectrometry-Based Strategies for Protein Disulfide Bond Identification. Rev. Anal. Chem. 2013, 32, 257–268. [Google Scholar] [CrossRef]
- Baumann, W.K.; Bizzozero, S.A.; Dutler, H. Specificity of α-Chymotrypsin. Dipeptide Substrates. FEBS Lett. 1970, 8, 257–260. [Google Scholar] [CrossRef] [Green Version]
- Keil, B. Specificity of Proteolysis; Springer: Berlin/Heidelberg, Germany, 1992; ISBN 9783642483820. [Google Scholar]
- Yuan, D.-D.; Han, Y.-H.; Wang, C.-G.; Chi, C.-W. From the Identification of Gene Organization of α Conotoxins to the Cloning of Novel Toxins. Toxicon 2007, 49, 1135–1149. [Google Scholar] [CrossRef] [PubMed]
- Kohn, A.J. Piscivorous gastropods of the genus conus. Proc. Natl. Acad. Sci. USA 1956, 42, 168–171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olivera, B.M.; Seger, J.; Horvath, M.P.; Fedosov, A.E. Prey-Capture Strategies of Fish-Hunting Cone Snails: Behavior, Neurobiology and Evolution. Brain Behav. Evol. 2015, 86, 58–74. [Google Scholar] [CrossRef] [Green Version]
- Rivera-Ortiz, J.A.; Cano, H.; Marí, F. Intraspecies Variability and Conopeptide Profiling of the Injected Venom of Conus Ermineus. Peptides 2011, 32, 306–316. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez, A.M.; Dutertre, S.; Lewis, R.J.; Marí, F. Intraspecific Variations in Conus Purpurascens Injected Venom Using LC/MALDI-TOF-MS and LC-ESI-TripleTOF-MS. Anal. Bioanal. Chem. 2015, 407, 6105–6116. [Google Scholar] [CrossRef]
- Le Gall, F.; Favreau, P.; Benoit, E.; Mattei, C.; Bouet, F.; Menou, J.-L.; Ménez, A.; Letourneux, Y.; Molgó, J. A New Conotoxin Isolated from Conus Consors Venom Acting Selectively on Axons and Motor Nerve Terminals through a Na+-Dependent Mechanism. Eur. J. Neurosci. 1999, 11, 3134–3142. [Google Scholar] [CrossRef]
- Hocking, H.G.; Gerwig, G.J.; Dutertre, S.; Violette, A.; Favreau, P.; Stöcklin, R.; Kamerling, J.P.; Boelens, R. Structure of the O-Glycosylated Conopeptide CcTx from Conus Consors Venom. Chem. Eur. J. 2013, 19, 870–879. [Google Scholar] [CrossRef]
- Lampe, R.A.; Lo, M.M.S.; Keith, R.A.; Horn, M.B.; McLane, M.W.; Herman, J.L.; Spreen, R.C. Effects of Site-Specific Acetylation on Omega.-Conotoxin GVIA Binding and Function. Biochemistry 1993, 32, 3255–3260. [Google Scholar] [CrossRef]
- Papineni, R.V.L.; Sanchez, J.U.; Baksi, K.; Willcockson, I.U.; Pedersen, S.E. Site-Specific Charge Interactions of α-Conotoxin MI with the Nicotinic Acetylcholine Receptor. J. Biol. Chem. 2001, 276, 23589–23598. [Google Scholar] [CrossRef]
- Himaya, S.W.A.; Jin, A.-H.; Hamilton, B.; Rai, S.K.; Alewood, P.; Lewis, R.J. Venom Duct Origins of Prey Capture and Defensive Conotoxins in Piscivorous Conus Striatus. Sci. Rep. 2021, 11, 13282. [Google Scholar] [CrossRef] [PubMed]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saintmont, F.; Cazals, G.; Bich, C.; Dutertre, S. Proteomic Analysis of the Predatory Venom of Conus striatus Reveals Novel and Population-Specific κA-Conotoxin SIVC. Toxins 2022, 14, 799. https://doi.org/10.3390/toxins14110799
Saintmont F, Cazals G, Bich C, Dutertre S. Proteomic Analysis of the Predatory Venom of Conus striatus Reveals Novel and Population-Specific κA-Conotoxin SIVC. Toxins. 2022; 14(11):799. https://doi.org/10.3390/toxins14110799
Chicago/Turabian StyleSaintmont, Fabrice, Guillaume Cazals, Claudia Bich, and Sebastien Dutertre. 2022. "Proteomic Analysis of the Predatory Venom of Conus striatus Reveals Novel and Population-Specific κA-Conotoxin SIVC" Toxins 14, no. 11: 799. https://doi.org/10.3390/toxins14110799