Seasonal Variability of the Airborne Eukaryotic Community Structure at a Coastal Site of the Central Mediterranean
Abstract
:1. Introduction
2. Results and Discussion
2.1. Mass Concentrations and Meteorological Parameters
2.2. Eukaryotic Community Structure at the Kingdom Level
2.3. Viridiplantae and Fungi Community Structures at the Phylum Level
2.4. Richness, Diversity, and Seasonal Dependence of Viridiplantae and Fungi Genera
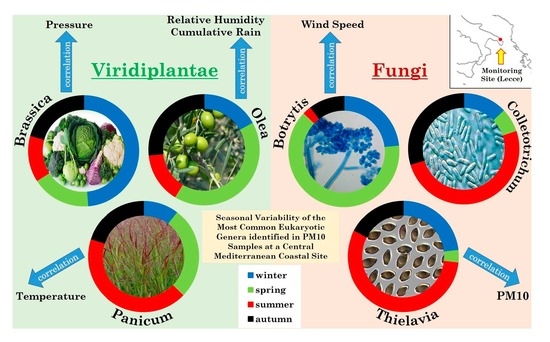
2.4.1. Overview of Streptophyta Genera in the PM10 Samples
2.4.2. Overview of Ascomycota and Basidiomycota Genus Communities in PM10 Samples
2.5. PCoA Analyses of Streptophyta and Ascomycota/Basidiomycota Genera in PM10 Samples
2.6. Relationships among Streptophyta and Ascomycota/Basidiomycota Genera, and with Meteorological Parameters and PM10 Mass Concentrations, by Spearman’s Correlation Coefficients
2.7. Potential Pathogenic Fungi and Plant-Derived Allergens in PM10 Samples
3. Summary and Conclusions
4. Material and Methods
4.1. Sampling Site, PM10 Sample Collection, and Meteorological Data
4.2. Long-Range Transported Air Masses at the Study Site
4.3. DNA Extraction and 18SrRNA Gene High-Throughput Sequencing
4.4. Statistical Analyses and Software
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gusareva, E.S.; Acerbi, E.; Lau, K.J.X.; Luhung, I.; Premkrishnan, B.N.V.; Kolundžija, S.; Purbojati, R.W.; Wong, A.; Houghton, J.N.I.; Miller, D.; et al. Microbial communities in the tropical air ecosystem follow a precise diel cycle. Proc. Natl. Acad. Sci. USA 2019, 116, 23299–23308. [Google Scholar] [CrossRef] [Green Version]
- Womack, A.M.; Bohannan, B.J.M.; Green, J.L. Biodiversity and biogeography of the atmosphere. Philos. Trans. R. Soc. B. Biol. Sci. 2010, 365, 3645–3653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Núñez, A.; García, A.M.; Moreno, D.A.; Guantes, R. Seasonal changes dominate long-term variability of the urban air microbiome across space and time. Environ. Int. 2021, 150, 106423. [Google Scholar] [CrossRef]
- Romano, S.; Becagli, S.; Lucarelli, F.; Rispoli, G.; Perrone, M. Airborne bacteria structure and chemical composition relationships in winter and spring PM10 samples over southeastern Italy. Sci. Total Environ. 2020, 730, 138899. [Google Scholar] [CrossRef]
- Maki, T.; Lee, K.C.; Kawai, K.; Onishi, K.; Hong, C.S.; Kurosaki, Y.; Shinoda, M.; Kai, K.; Iwasaka, Y.; Archer, S.D.J.; et al. Aeolian Dispersal of Bacteria Associated With Desert Dust and Anthropogenic Particles Over Continental and Oceanic Surfaces. J. Geophys. Res. Atmos. 2019, 124, 5579–5588. [Google Scholar] [CrossRef]
- Falkowski, P.G.; Fenchel, T.; Delong, E.F. The Microbial Engines That Drive Earth’s Biogeochemical Cycles. Science 2008, 320, 1034–1039. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez-Martin, C. Airborne Infectious Microorganisms. In Encyclopedia of Microbiology; Schmidt, T.M., Ed.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 52–60. [Google Scholar] [CrossRef]
- Jones, R.M.; Brosseau, L.M. Aerosol Transmission of Infectious Disease. J. Occup. Environ. Med. 2015, 57, 501–508. [Google Scholar] [CrossRef]
- Bourdrel, T.; Annesi-Maesano, I.; Alahmad, B.; Maesano, C.N.; Bind, M.-A. The impact of outdoor air pollution on COVID-19: A review of evidence from in vitro, animal, and human studies. Eur. Respir. Rev. 2021, 30, 200242. [Google Scholar] [CrossRef]
- Chen, X.; Kumari, D.; Achal, V. A Review on Airborne Microbes: The Characteristics of Sources, Pathogenicity and Geography. Atmosphere 2020, 11, 919. [Google Scholar] [CrossRef]
- Bowers, R.M.; Clements, N.; Emerson, J.; Wiedinmyer, C.; Hannigan, M.; Fierer, N. Seasonal Variability in Bacterial and Fungal Diversity of the Near-Surface Atmosphere. Environ. Sci. Technol. 2013, 47, 12097–12106. [Google Scholar] [CrossRef] [PubMed]
- Gandolfi, I.; Bertolini, V.; Ambrosini, R.; Bestetti, G.; Franzetti, A. Unravelling the bacterial diversity in the atmosphere. Appl. Microbiol. Biotechnol. 2013, 97, 4727–4736. [Google Scholar] [CrossRef]
- Aziz, A.A.; Lee, K.; Park, B.; Park, H.; Park, K.; Choi, I.-G.; Chang, I.S. Comparative study of the airborne microbial communities and their functional composition in fine particulate matter (PM2.5) under non-extreme and extreme PM2.5 conditions. Atmos. Environ. 2018, 194, 82–92. [Google Scholar] [CrossRef]
- Song, H.; Crawford, I.; Lloyd, J.; Robinson, C.; Boothman, C.; Bower, K.; Gallagher, M.; Allen, G.; Topping, D. Airborne Bacterial and Eukaryotic Community Structure across the United Kingdom Revealed by High-Throughput Sequencing. Atmosphere 2020, 11, 802. [Google Scholar] [CrossRef]
- Ruiz-Gil, T.; Acuña, J.J.; Fujiyoshi, S.; Tanaka, D.; Noda, J.; Maruyama, F.; Jorquera, M.A. Airborne bacterial communities of outdoor environments and their associated influencing factors. Environ. Int. 2020, 145, 106156. [Google Scholar] [CrossRef]
- Calderón-Ezquerro, M.D.C.; Serrano-Silva, N.; Brunner-Mendoza, C. Aerobiological study of bacterial and fungal community composition in the atmosphere of Mexico City throughout an annual cycle. Environ. Pollut. 2021, 278, 116858. [Google Scholar] [CrossRef]
- Perrone, M.; Becagli, S.; Orza, J.A.G.; Vecchi, R.; Dinoi, A.; Udisti, R.; Cabello, M. The impact of long-range-transport on PM1 and PM2.5 at a Central Mediterranean site. Atmos. Environ. 2013, 71, 176–186. [Google Scholar] [CrossRef]
- Romano, S.; Di Salvo, M.; Rispoli, G.; Alifano, P.; Perrone, M.R.; Talà, A. Airborne bacteria in the Central Mediterranean: Structure and role of meteorology and air mass transport. Sci. Total Environ. 2019, 697, 134020. [Google Scholar] [CrossRef]
- Romano, S.; Fragola, M.; Alifano, P.; Perrone, M.; Talà, A. Potential Human and Plant Pathogenic Species in Airborne PM10 Samples and Relationships with Chemical Components and Meteorological Parameters. Atmosphere 2021, 12, 654. [Google Scholar] [CrossRef]
- Pietrogrande, M.C.; Perrone, M.R.; Manarini, F.; Romano, S.; Udisti, R.; Becagli, S. PM10 oxidative potential at a Central Mediterranean Site: Association with chemical composition and meteorological parameters. Atmos. Environ. 2018, 188, 97–111. [Google Scholar] [CrossRef]
- Perrone, M.; Romano, S.; Genga, A.; Paladini, F. Integration of optical and chemical parameters to improve the particulate matter characterization. Atmos. Res. 2018, 205, 93–106. [Google Scholar] [CrossRef]
- Perrone, M.R.; Romano, S.; Orza, J. Particle optical properties at a Central Mediterranean site: Impact of advection routes and local meteorology. Atmos. Res. 2014, 145–146, 152–167. [Google Scholar] [CrossRef]
- O’Neill, M.A.; Darvill, A.G.; Etzler, M.E.; Mohnen, D.; Perez, S. Viridiplantae and Algae. In Essentials of Glycobiology [Internet], 3rd ed.; Varki, A., Cummings, R.D., Esko, J.D., et al., Eds.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2017; Chapter 24. [Google Scholar]
- Banchi, E.; Ametrano, C.G.; Tordoni, E.; Stanković, D.; Ongaro, S.; Tretiach, M.; Pallavicini, A.; Muggia, L.; Verardo, P.; Tassan, F.; et al. Environmental DNA assessment of airborne plant and fungal seasonal diversity. Sci. Total Environ. 2020, 738, 140249. [Google Scholar] [CrossRef] [PubMed]
- Núñez, A.; de Paz, G.A.; Rastrojo, A.; Ferencova, Z.; Gutiérrez-Bustillo, A.M.; Alcamí, A.; Moreno, D.A.; Guantes, R. Temporal patterns of variability for prokaryotic and eukaryotic diversity in the urban air of Madrid (Spain). Atmos. Environ. 2019, 217, 116972. [Google Scholar] [CrossRef]
- Han, B.; Weiss, L.M. Microsporidia: Obligate Intracellular Pathogens Within the Fungal Kingdom. In The Fungal Kingdom; ASM Press: Washington, DC, USA, 2017; pp. 97–113. [Google Scholar] [CrossRef] [Green Version]
- Du, P.; Du, R.; Ren, W.; Lu, Z.; Zhang, Y.; Fu, P. Variations of bacteria and fungi in PM2.5 in Beijing, China. Atmos. Environ. 2018, 172, 55–64. [Google Scholar] [CrossRef]
- Banchi, E.; Pallavicini, A.; Muggia, L. Relevance of plant and fungal DNA metabarcoding in aerobiology. Aerobiologia 2019, 36, 9–23. [Google Scholar] [CrossRef]
- Shi, C.-F.; Zhang, K.-H.; Chai, C.-Y.; Yan, Z.-L.; Hui, F.-L. Diversity of the genus Sugiyamaella and description of two new species from rotting wood in China. MycoKeys 2021, 77, 27–39. [Google Scholar] [CrossRef]
- Ramette, A.N. Multivariate analyses in microbial ecology. FEMS Microbiol. Ecol. 2007, 62, 142–160. [Google Scholar] [CrossRef] [Green Version]
- Janssen, R.H.H.; Heald, C.L.; Steiner, A.L.; Perring, A.E.; Huffman, J.A.; Robinson, E.S.; Twohy, C.H.; Ziemba, L.D. Drivers of the fungal spore bioaerosol budget: Observational analysis and global modeling. Atmos. Chem. Phys. Discuss. 2021, 21, 4381–4401. [Google Scholar] [CrossRef]
- Brown, E.; McTaggart, L.R.; Low, D.E.; Richardson, S.E. Effective method for the heat inactivation of Blastomyces dermatitidis. Med. Mycol. 2014, 52, 766–769. [Google Scholar] [CrossRef] [Green Version]
- Singh, N.K.; Wood, J.M.; Karouia, F.; Venkateswaran, K. Succession and persistence of microbial communities and antimicrobial resistance genes associated with International Space Station environmental surfaces. Microbiome 2018, 6, 204. [Google Scholar] [CrossRef] [Green Version]
- Pfliegler, W.P.; Pócsi, I.; Győri, Z.; Pusztahelyi, T. The Aspergilli and Their Mycotoxins: Metabolic Interactions With Plants and the Soil Biota. Front. Microbiol. 2020, 10, 2921. [Google Scholar] [CrossRef] [Green Version]
- Foley, K.; Fazio, G.; Jensen, A.B.; Hughes, W.O. The distribution of Aspergillus spp. opportunistic parasites in hives and their pathogenicity to honey bees. Vet. Microbiol. 2014, 169, 203–210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paulussen, C.; Hallsworth, J.E.; Álvarez-Pérez, S.; Nierman, W.C.; Hamill, P.G.; Blain, D.; Rediers, H.; Lievens, B. Ecology of aspergillosis: Insights into the pathogenic potency of Aspergillus fumigatus and some other Aspergillus species. Microb. Biotechnol. 2016, 10, 296–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guinea, J.; Peláez, T.; Alcalá, L.; Bouza, E. Outdoor environmental levels of Aspergillus spp. conidia over a wide geographical area. Med. Mycol. 2006, 44, 349–356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- R, H.P.; Singh, R.K.; Akila, M.; Ravikrishna, R.; Verma, R.; Gunthe, S.S. Seasonal variation of the dominant allergenic fungal aerosols–One year study from southern Indian region. Sci. Rep. 2017, 7, 11171. [Google Scholar] [CrossRef] [Green Version]
- Aydoğdu, M.; Kurbetli, I.; Canpolat, S. Batı Akdeniz Bölgesi’nde Enginar baş çürüklüğü (Botrytis cinerea Pers.) hastalığının yaygınlığı ve üretime etkisinin belirlenmesi. Bitki Koruma Bülteni 2020, 60, 21–29. [Google Scholar] [CrossRef]
- Williamson, B.; Tudzynski, B.; Tudzynski, P.; van Kan, J. Botrytis cinerea: The cause of grey mould disease. Mol. Plant Pathol. 2007, 8, 561–580. [Google Scholar] [CrossRef]
- Jurgensen, C.W.; Madsen, A.M. Exposure to the airborne mould Botrytis and its health effects. Ann. Agric. Environ. Med. 2009, 16, 183–196. [Google Scholar]
- Hashimoto, S.; Tanaka, E.; Ueyama, M.; Terada, S.; Inao, T.; Kaji, Y.; Yasuda, T.; Hajiro, T.; Nakagawa, T.; Noma, S.; et al. A case report of pulmonary Botrytis sp. infection in an apparently healthy individual. BMC Infect. Dis. 2019, 19, 1–8. [Google Scholar] [CrossRef]
- Monteil, C.; Bardin, M.; E Morris, C. Features of air masses associated with the deposition of Pseudomonas syringae and Botrytis cinerea by rain and snowfall. ISME J. 2014, 8, 2290–2304. [Google Scholar] [CrossRef] [Green Version]
- Elad, Y.; Williamson, B.; Tudzynski, P.; Delen, N. Botrytis spp. and Diseases They Cause in Agricultural Systems–An Introduction. In Botrytis: Biology, Pathology and Control; Springer: Dordrecht, The Netherlands, 2007; pp. 1–8. [Google Scholar]
- Blanco, C.; De Santos, B.L.; Romero, F. Relationship between Concentrations of Botrytis Cinerea Conidia in Air, Environmental Conditions, and the Incidence of Grey Mould in Strawberry Flowers and Fruits. Eur. J. Plant Pathol. 2006, 114, 415–425. [Google Scholar] [CrossRef]
- Summerell, B.A. Resolving Fusarium: Current Status of the Genus. Annu. Rev. Phytopathol. 2019, 57, 323–339. [Google Scholar] [CrossRef]
- Hof, H. The Medical Relevance of Fusarium spp. J. Fungi 2020, 6, 117. [Google Scholar] [CrossRef] [PubMed]
- Gupta, G.; Fries, B.C. Variability of phenotypic traits in Cryptococcus varieties and species and the resulting implications for pathogenesis. Futur. Microbiol. 2010, 5, 775–787. [Google Scholar] [CrossRef] [Green Version]
- Pal, M. Cryptococcus gattii: An emerging global mycotic pathogen of humans and animals. J. Mycopathol. Res. 2014, 52, 1–7. [Google Scholar]
- Da Silva, L.L.; Moreno, H.L.A.; Correia, H.L.N.; Santana, M.; De Queiroz, M.V. Colletotrichum: Species complexes, lifestyle, and peculiarities of some sources of genetic variability. Appl. Microbiol. Biotechnol. 2020, 104, 1891–1904. [Google Scholar] [CrossRef] [PubMed]
- Perelló, A.; Gasso, M.M.A.; Lovisolo, M. Biology and histopathology of Ustilago filiformis (=U. longissima), a causal agent of leaf stripe smut of Glyceria multiflora. J. Plant Prot. Res. 2015, 55, 429–437. [Google Scholar] [CrossRef]
- Joshi, R. A Review on Colletotrichum spp. Virulence mechanism against host plant defensive factors. J. Med. Plants Stud. 2018, 6, 64–67. [Google Scholar] [CrossRef]
- Valle-Aguirre, G.; Valle, M.G.V.-D.; Corona-Rangel, M.L.; Amora-Lazcano, E.; Hernández-Lauzardo, A.N. First aeromycological study in an avocado agroecosystem in Mexico. Aerobiologia 2016, 32, 657–667. [Google Scholar] [CrossRef]
- D’Amato, G. Effects of climatic changes and urban air pollution on the rising trends of respiratory allergy and asthma. Multidiscip. Respir. Med. 2011, 6, 28–37. [Google Scholar] [CrossRef] [Green Version]
- Kadam, K.; Karbhal, R.; Jayaraman, V.K.; Sawant, S.; Kulkarni-Kale, U. AllerBase: A comprehensive allergen knowledgebase. Database 2017, 2017. [Google Scholar] [CrossRef]
- Radauer, C.; Breiteneder, H. Allergen databases—A critical evaluation. Allergy 2019, 74, 2057–2060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burton, N.C.; Grinshpun, S.A.; Reponen, T. Physical Collection Efficiency of Filter Materials for Bacteria and Viruses. Ann. Occup. Hyg. 2006, 51, 143–151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mykytczuk, N.C.S.; Wilhelm, R.C.; Whyte, L.G. Planococcus halocryophilus sp. nov., an extreme sub-zero species from high Arctic permafrost. Int. J. Syst. Evol. Microbiol. 2012, 62, 1937–1944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Basart, S.; Pérez, C.; Cuevas, E.; Baldasano, J.M.; Gobbi, G.P. Aerosol characterization in Northern Africa, Northeastern Atlantic, Mediterranean Basin and Middle East from direct-sun AERONET observations. Atmos. Chem. Phys. Discuss. 2009, 9, 8265–8282. [Google Scholar] [CrossRef] [Green Version]
- Perrone, M.R.; Romano, S.; Orza, J.A.G. Columnar and ground-level aerosol optical properties: Sensitivity to the transboundary pollution, daily and weekly patterns, and relationships. Environ. Sci. Pollut. Res. 2015, 22, 16570–16589. [Google Scholar] [CrossRef]
- Perrone, M.R.; Vecchi, R.; Romano, S.; Becagli, S.; Traversi, R.; Paladini, F. Weekly cycle assessment of PM mass concentrations and sources, and impacts on temperature and wind speed in Southern Italy. Atmos. Res. 2019, 218, 129–144. [Google Scholar] [CrossRef]
- Mallet, M.; Dulac, F.; Formenti, P.; Nabat, P.; Sciare, J.; Roberts, G.; Pelon, J.; Ancellet, G.; Tanré, D.; Parol, F.; et al. Overview of the Chemistry-Aerosol Mediterranean Experiment/Aerosol Direct Radiative Forcing on the Mediterranean Climate (ChArMEx/ADRIMED) summer 2013 campaign. Atmos. Chem. Phys. 2016, 16, 455–504. [Google Scholar] [CrossRef] [Green Version]
- Stein, A.F.; Draxler, R.R.; Rolph, G.D.; Stunder, B.J.B.; Cohen, M.D.; Ngan, F. NOAA’s HYSPLIT Atmospheric Transport and Dispersion Modeling System. Bull. Am. Meteorol. Soc. 2015, 96, 2059–2077. [Google Scholar] [CrossRef]
- Haustein, K.; Pérez, C.; Baldasano, J.M.; Jorba, O.; Basart, S.; Miller, R.L.; Janjić, Z.; Black, T.; Nickovic, S.; Todd, M.C.; et al. Atmospheric dust modeling from meso to global scales with the online NMMB/BSC-Dust model–Part 2: Experimental campaigns in Northern Africa. Atmos. Chem. Phys. Discuss. 2012, 12, 2933–2958. [Google Scholar] [CrossRef] [Green Version]
- Després, V.R.; Huffman, J.A.; Burrows, S.M.; Hoose, C.; Safatov, A.S.; Buryak, G.; Fröhlich-Nowoisky, J.; Elbert, W.; Andreae, M.O.; Pöschl, U.; et al. Primary biological aerosol particles in the atmosphere: A review. Tellus B. Chem. Phys. Meteorol. 2012, 64, 15598. [Google Scholar] [CrossRef] [Green Version]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar] [CrossRef]
- Wood, D.E.; Salzberg, S.L. Kraken: Ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014, 15, R46. [Google Scholar] [CrossRef] [Green Version]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shannon, C.E. A mathematical theory of communication. Bell Labs Tech. J. 1948, 27, 379–423. [Google Scholar] [CrossRef] [Green Version]
- Simpson, E. Measurement of diversity. Nature 1949, 163, 688. [Google Scholar] [CrossRef]
- Haegeman, B.; Hamelin, J.; Moriarty, J.; Neal, P.; Dushoff, J.; Weitz, J.S. Robust estimation of microbial diversity in theory and in practice. ISME J. 2013, 7, 1092–1101. [Google Scholar] [CrossRef]
- Chernov, T.I.; Tkhakakhova, A.K.; Kutovaya, O.V. Assessment of diversity indices for the characterization of the soil prokaryotic community by metagenomic analysis. Eurasian Soil Sci. 2015, 48, 410–415. [Google Scholar] [CrossRef]
- Escobar-Zepeda, A.; De León, A.V.-P.; Sanchez-Flores, A. The Road to Metagenomics: From Microbiology to DNA Sequencing Technologies and Bioinformatics. Front. Genet. 2015, 6, 348. [Google Scholar] [CrossRef] [Green Version]
- Kim, B.-R.; Shin, J.; Guevarra, R.B.; Lee, J.H.; Kim, D.W.; Seol, K.-H.; Lee, J.-H.; Kim, H.B.; Isaacson, R.E. Deciphering Diversity Indices for a Better Understanding of Microbial Communities. J. Microbiol. Biotechnol. 2017, 27, 2089–2093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krebs, C.J. Species diversity measures. In Ecological Methodology; Krebs, C.J., Ed.; University of British Columbia: Vancouver, BC, Canada, 2014; pp. 532–593. [Google Scholar]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [Green Version]
- Magurran, A.E. An index of diversity. In Measuring Biological Diversity; Blackwell Science: Oxford, UK, 2004; Chapter 4. [Google Scholar]
- Ricotta, C.; Podani, J. On some properties of the Bray-Curtis dissimilarity and their ecological meaning. Ecol. Complex. 2017, 31, 201–205. [Google Scholar] [CrossRef]
- Hammer, O.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar]
- Hervé, M.R.; Nicolè, F.; Le Cao, K.-A. Multivariate Analysis of Multiple Datasets: A Practical Guide for Chemical Ecology. J. Chem. Ecol. 2018, 44, 215–234. [Google Scholar] [CrossRef] [PubMed]
- Legendre, P.; Anderson, M.J. Distance-Based Redundancy Analysis: Testing Multispecies Responses in Multifactorial Ecological Experiments. Ecol. Monogr. 1999, 69, 1–24. [Google Scholar] [CrossRef]
- Jones, D.L. Fathom Toolbox for MATLAB: Software for Multivariate Ecological and Oceanographic Data Analysis; College of Marine Science, University of South Florida: St. Petersburg, FL, USA, 2017; Available online: https://www.marine.usf.edu/research/matlab-resources/ (accessed on 17 May 2021).







| Season | PM10 | T | RH | P | CR | WD | WS |
|---|---|---|---|---|---|---|---|
| (µg m−3) | (°C) | (%) | (mbar) | (mm) | (deg) | (ms−1) | |
| Winter (mean ± SD) | 25 ± 15 | 8.7 ± 1.6 | 65 ± 13 | 1012.4 ± 11.5 | 39.1 | 341 ± 31 | 2.4 ± 1.4 |
| Spring (mean ± SD) | 20 ± 5 | 16.9 ± 3.4 | 72 ± 7 | 1011.4 ± 4.2 | 29.2 | 126 ± 59 | 2.2 ± 0.9 |
| Summer (mean ± SD) | 24 ± 4 | 26.1 ± 1.1 | 57 ± 5 | 1009.5 ± 3.3 | 0.0 | 348 ± 34 | 2.0 ± 0.9 |
| Autumn (mean ± SD) | 22 ± 11 | 12.7 ± 4.6 | 76 ± 9 | 1013.9 ± 4.6 | 32.6 | 329 ± 8 | 1.5 ± 1.0 |
| Sample | Viridiplantae | Fungi | ||||||
|---|---|---|---|---|---|---|---|---|
| n° OTUs | n° Genera | At Genus Level | n° OTUs | n° Genera | At Genus Level | |||
| Shannon Index (H) | Simpson Index (D) | Shannon Index (H) | Simpson Index (D) | |||||
| S1 | 143 | 42 | 2.33 | 0.14 | 77 | 19 | 2.26 | 0.13 |
| S2 | 149 | 43 | 2.33 | 0.14 | 66 | 14 | 1.90 | 0.18 |
| S3 | 142 | 42 | 1.98 | 0.21 | 77 | 18 | 2.02 | 0.18 |
| S4 | 134 | 39 | 2.04 | 0.19 | 67 | 16 | 2.00 | 0.21 |
| S5 | 141 | 41 | 1.94 | 0.23 | 76 | 18 | 2.02 | 0.21 |
| S6 | 140 | 39 | 2.28 | 0.20 | 70 | 16 | 2.11 | 0.17 |
| S7 | 142 | 40 | 0.91 | 0.69 | 58 | 13 | 2.02 | 0.17 |
| S8 | 142 | 42 | 1.04 | 0.65 | 60 | 14 | 2.18 | 0.14 |
| S9 | 150 | 44 | 2.08 | 0.20 | 65 | 15 | 1.81 | 0.24 |
| S10 | 140 | 40 | 2.34 | 0.15 | 73 | 17 | 1.42 | 0.37 |
| S11 | 144 | 42 | 2.04 | 0.22 | 75 | 18 | 1.06 | 0.52 |
| S12 | 140 | 41 | 2.05 | 0.21 | 76 | 18 | 1.41 | 0.42 |
| S13 | 145 | 43 | 2.13 | 0.18 | 73 | 17 | 1.10 | 0.57 |
| S14 | 140 | 41 | 1.77 | 0.29 | 67 | 16 | 1.61 | 0.31 |
| S15 | 145 | 43 | 1.80 | 0.26 | 76 | 18 | 1.08 | 0.55 |
| S16 | 144 | 43 | 1.71 | 0.35 | 72 | 17 | 1.67 | 0.28 |
| S17 | 140 | 41 | 2.43 | 0.12 | 69 | 16 | 1.74 | 0.24 |
| S18 | 152 | 45 | 2.17 | 0.18 | 76 | 18 | 1.95 | 0.20 |
| S19 | 139 | 42 | 2.01 | 0.25 | 68 | 16 | 1.73 | 0.23 |
| S20 | 150 | 44 | 2.25 | 0.15 | 71 | 16 | 1.80 | 0.23 |
| S21 | 147 | 44 | 2.09 | 0.17 | 77 | 18 | 2.27 | 0.12 |
| S22 | 137 | 40 | 2.07 | 0.20 | 76 | 18 | 1.68 | 0.26 |
| S23 | 130 | 37 | 2.29 | 0.14 | 73 | 17 | 1.30 | 0.43 |
| S24 | 150 | 44 | 2.26 | 0.15 | 72 | 16 | 1.83 | 0.23 |
| S25 | 143 | 42 | 2.10 | 0.17 | 75 | 18 | 1.93 | 0.20 |
| S26 | 128 | 37 | 1.97 | 0.22 | 69 | 16 | 1.44 | 0.40 |
| S27 | 140 | 41 | 2.13 | 0.17 | 73 | 17 | 2.33 | 0.11 |
| S28 | 106 | 30 | 1.41 | 0.44 | 61 | 14 | 1.52 | 0.27 |
| S29 | 142 | 42 | 2.25 | 0.15 | 74 | 17 | 2.33 | 0.12 |
| S30 | 128 | 37 | 1.83 | 0.24 | 67 | 17 | 1.46 | 0.37 |
| S31 | 146 | 43 | 2.08 | 0.17 | 76 | 18 | 2.18 | 0.15 |
| S32 | 146 | 43 | 1.89 | 0.25 | 69 | 16 | 2.07 | 0.18 |
| S33 | 131 | 39 | 2.43 | 0.13 | 65 | 15 | 1.98 | 0.18 |
| S34 | 130 | 37 | 1.87 | 0.25 | 66 | 14 | 2.15 | 0.16 |
| S35 | 146 | 43 | 2.41 | 0.12 | 75 | 18 | 2.30 | 0.12 |
| S36 | 143 | 43 | 2.31 | 0.14 | 66 | 16 | 2.01 | 0.20 |
| S37 | 146 | 43 | 1.95 | 0.23 | 74 | 18 | 1.83 | 0.22 |
| Streptophyta Genera | Positive Correlations | Fungi Phyla | Fungi Genera | Positive Correlations |
|---|---|---|---|---|
| Brassica (BRA) | ASP (0.38), SCHE (0.38), P (0.33) | ASCOMYCOTA | Botrytis (BOT) | CRY (0.57), WS (0.39) |
| Olea (OLE) | SES (0.37), RH (0.39), CR (0.36) | Colletotrichum (COL) | THI (0.41), SUG (0.45), NEU (0.48) | |
| Panicum (PAN) | UST (0.43), T (0.50) | Thielavia (THI) | COL (0.41), SUG (0.47), ASP (0.40), PM10 (0.34) | |
| Beta (BET) | PHY (0.41), NIC (0.33), CIC (0.66), COL (0.36), NEU (0.34), SCHE (0.53), MAL (0.40), CR (0.49) | Sugiyamaella (SUG) | THI (0.47), MAL (0.38) | |
| Physcomitrella (PHY) | BET (0.41), CIC (0.40), COL (0.60), THI (0.44), SUG (0.46), ASP (0.37), NEU (0.60) | Aspergillus (ASP) | THI (0.40), POC (0.35), NEU (0.44), FUS (0.73) | |
| Gossypium (GOS) | SES (0.34), PM10 (0.33) | Pochonia (POC) | ASP (0.35), SCHE (0.47), FUS (0.45), CRY (0.39), CR (0.43), WS (0.39) | |
| Capsicum (CAP) | SES (0.45), BOT (0.51), POC (0.41) | Neurospora (NEU) | COL (0.48), ASP (0.44), FUS (0.43) | |
| Nicotiana (NIC) | BET (0.33), SES (0.62), CIC (0.48), MAL (0.36) | Scheffersomyces (SCHE) | POC (0.47), FUS (0.39), CRY (0.48), CR (0.60) | |
| Daucus (DAU) | NEU (0.35) | Fusarium (FUS) | ASP (0.73), POC (0.45), NEU (0.43), SCHE (0.39) | |
| Sesamum (SES) | OLE (0.37), GOS (0.34), CAP (0.45), NIC (0.62), BOT (0.36) | BASIDIOMYCOTA | Cryptococcus (CRY) | BOT (0.57), POC (0.39), SCHE (0.48) |
| Cicer (CIC) | BET (0.66), PHY (0.40), NIC (0.48), SUG (0.35) | Ustilago (UST) | T (0.37) | |
| Lupinus (LUP) | ASP (0.38), UST (0.47) | Malassezia (MAL) | SUG (0.38), RH (0.36) | |
| T | CR | RH (0.36) | ||
| RH | CR (0.36) | WS | ||
| PM10 | P (0.33) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fragola, M.; Perrone, M.R.; Alifano, P.; Talà, A.; Romano, S. Seasonal Variability of the Airborne Eukaryotic Community Structure at a Coastal Site of the Central Mediterranean. Toxins 2021, 13, 518. https://doi.org/10.3390/toxins13080518
Fragola M, Perrone MR, Alifano P, Talà A, Romano S. Seasonal Variability of the Airborne Eukaryotic Community Structure at a Coastal Site of the Central Mediterranean. Toxins. 2021; 13(8):518. https://doi.org/10.3390/toxins13080518
Chicago/Turabian StyleFragola, Mattia, Maria Rita Perrone, Pietro Alifano, Adelfia Talà, and Salvatore Romano. 2021. "Seasonal Variability of the Airborne Eukaryotic Community Structure at a Coastal Site of the Central Mediterranean" Toxins 13, no. 8: 518. https://doi.org/10.3390/toxins13080518