Bothrops Jararaca Snake Venom Modulates Key Cancer-Related Proteins in Breast Tumor Cell Lines
Abstract
:1. Introduction
2. Results
2.1. The Cytotoxicity of B. jararaca Snake Venom in MCF7 and MDA-MB-231 Cells
2.2. Optical Microscopy Analysis of MCF7 and MDA-MB-231 Cells under B. jararaca Venom Treatment
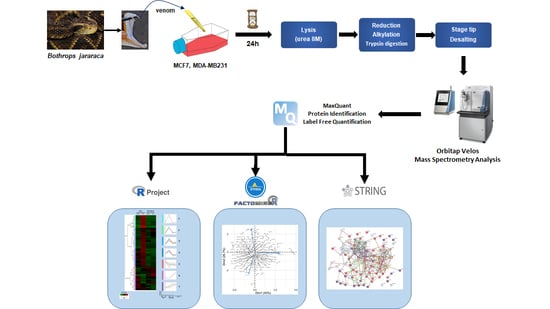
2.3. Mass Spectrometry-Based Proteomics of MCF7 and MDA-MB-231 Cells Treated with B. jararaca Venom
2.4. Semi-Quantitative Proteomics Analysis: MCF7 and MDA-MB-231 Cell Line Protein Abundance Variation
2.5. Hierarchical Clustering Analysis
2.6. Principal Component Analysis
2.7. Gene Ontology Functional Analysis
2.8. Protein–Protein Interaction Analysis
2.9. Exclusive Proteins
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Cell Culture and Maintenance
5.2. Bothrops jararaca Venom
5.3. Cell Viability Assay
5.4. Cell Treatment with B. jararaca Venom
5.5. Sample Preparation for Proteomic Analysis
5.6. Mass Spectrometry Analysis
5.7. Proteome Functional and Enrichment Analysis
5.8. Semi-Quantitative Proteomics Analysis
5.9. Principal Component Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brazilian Ministry of Health. Accidents at Work by Venomous Animals among Workers at the Countryside, Forest, and Water in Brazil from 2007 to 2017 (Port). 2019; Volume 50. Available online: https://portalarquivos2.saude.gov.br/images/pdf/2019/marco/29/2018-059.pdf (accessed on 1 March 2021).
- Ribeiro, L.A.; Jorge, M.T. Bites by snakes in the genus Bothrops: A series of 3139 cases. Rev. Soc. Bras. Med. Trop. 1997, 30, 475–480. [Google Scholar] [CrossRef] [Green Version]
- Tanjoni, I.; Weinlich, R.; Della-Casa, M.S.; Clissa, P.B.; Saldanha-Gama, R.F.; de Freitas, M.S.; Barja-Fidalgo, C.; Amarante-Mendes, G.P.; Moura-da-Silva, A.M. Jararhagin, a snake venom metalloproteinase, induces a specialized form of apoptosis (anoikis) selective to endothelial cells. Apoptosis 2005, 10, 851–861. [Google Scholar] [CrossRef] [PubMed]
- Calvete, J.J.; Juarez, P.; Sanz, L. Snake venomics. Strategy and applications. J. Mass Spectrom. 2007, 42, 1405–1414. [Google Scholar] [CrossRef] [PubMed]
- Fox, J.W.; Serrano, S.M. Exploring snake venom proteomes: Multifaceted analyses for complex toxin mixtures. Proteomics 2008, 8, 909–920. [Google Scholar] [CrossRef] [PubMed]
- Doley, R.; Kini, R.M. Protein complexes in snake venom. Cell. Mol. Life Sci. 2009, 66, 2851–2871. [Google Scholar] [CrossRef]
- Serrano, S.M.; Shannon, J.D.; Wang, D.; Camargo, A.C.; Fox, J.W. A multifaceted analysis of viperid snake venoms by two-dimensional gel electrophoresis: An approach to understanding venom proteomics. Proteomics 2005, 5, 501–510. [Google Scholar] [CrossRef]
- Jin, H.; Varner, J. Integrins: Roles in cancer development and as treatment targets. Br. J. Cancer 2004, 90, 561–565. [Google Scholar] [CrossRef]
- White, J. Snake venoms and coagulopathy. Toxicon 2005, 45, 951–967. [Google Scholar] [CrossRef]
- Gay, C.; Sanz, L.; Calvete, J.J.; Pla, D. Snake Venomics and Antivenomics of Bothrops diporus, a Medically Important Pitviper in Northeastern Argentina. Toxins 2015, 8, 9. [Google Scholar] [CrossRef] [Green Version]
- Sanz, L.; Perez, A.; Quesada-Bernat, S.; Diniz-Sousa, R.; Calderon, L.A.; Soares, A.M.; Calvete, J.J.; Caldeira, C.A.S. Venomics and antivenomics of the poorly studied Brazil’s lancehead, Bothrops brazili (Hoge, 1954), from the Brazilian State of Para. J. Venom. Anim. Toxins Incl. Trop. Dis. 2020, 26, e20190103. [Google Scholar] [CrossRef]
- Tashima, A.K.; Zelanis, A.; Kitano, E.S.; Ianzer, D.; Melo, R.L.; Rioli, V.; Sant’anna, S.S.; Schenberg, A.C.; Camargo, A.C.; Serrano, S.M. Peptidomics of three Bothrops snake venoms: Insights into the molecular diversification of proteomes and peptidomes. Mol. Cell. Proteom. 2012, 11, 1245–1262. [Google Scholar] [CrossRef] [Green Version]
- Gren, E.C.K.; Kitano, E.S.; Andrade-Silva, D.; Iwai, L.K.; Reis, M.S.; Menezes, M.C.; Serrano, S.M.T. Comparative analysis of the high molecular mass subproteomes of eight Bothrops snake venoms. Comp. Biochem. Physiol. Part D Genom. Proteom. 2019, 30, 113–121. [Google Scholar] [CrossRef]
- Zelanis, A.; Tashima, A.K.; Pinto, A.F.; Paes Leme, A.F.; Stuginski, D.R.; Furtado, M.F.; Sherman, N.E.; Ho, P.L.; Fox, J.W.; Serrano, S.M. Bothrops jararaca venom proteome rearrangement upon neonate to adult transition. Proteomics 2011, 11, 4218–4228. [Google Scholar] [CrossRef]
- Farias, I.B.; Morais-Zani, K.; Serino-Silva, C.; Sant’Anna, S.S.; Rocha, M.; Grego, K.F.; Andrade-Silva, D.; Serrano, S.M.T.; Tanaka-Azevedo, A.M. Functional and proteomic comparison of Bothrops jararaca venom from captive specimens and the Brazilian Bothropic Reference Venom. J. Proteom. 2018, 174, 36–46. [Google Scholar] [CrossRef]
- Augusto-de-Oliveira, C.; Stuginski, D.R.; Kitano, E.S.; Andrade-Silva, D.; Liberato, T.; Fukushima, I.; Serrano, S.M.; Zelanis, A. Dynamic Rearrangement in Snake Venom Gland Proteome: Insights into Bothrops jararaca Intraspecific Venom Variation. J. Proteome Res. 2016, 15, 3752–3762. [Google Scholar] [CrossRef]
- Laing, G.D.; Clissa, P.B.; Theakston, R.D.; Moura-da-Silva, A.M.; Taylor, M.J. Inflammatory pathogenesis of snake venom metalloproteinase-induced skin necrosis. Eur. J. Immunol. 2003, 33, 3458–3463. [Google Scholar] [CrossRef] [Green Version]
- Azevedo-Marques, M.M.; Cupo, P.; Hering, S.E. Acidentes Por Animais PeÇonhentos: Serpentes PeÇonhentas. Medicina 2003, 36. [Google Scholar] [CrossRef] [Green Version]
- Bjarnason, J.B.; Fox, J.W. Hemorrhagic metalloproteinases from snake venoms. Pharmacol. Ther. 1994, 62, 325–372. [Google Scholar] [CrossRef]
- Cidade, D.A.; Simao, T.A.; Davila, A.M.; Wagner, G.; Junqueira-de-Azevedo, I.L.; Ho, P.L.; Bon, C.; Zingali, R.B.; Albano, R.M. Bothrops jararaca venom gland transcriptome: Analysis of the gene expression pattern. Toxicon 2006, 48, 437–461. [Google Scholar] [CrossRef]
- Pereira, L.M.; Messias, E.A.; Sorroche, B.P.; Oliveira, A.D.N.; Arantes, L.; de Carvalho, A.C.; Tanaka-Azevedo, A.M.; Grego, K.F.; Carvalho, A.L.; Melendez, M.E. In-depth transcriptome reveals the potential biotechnological application of Bothrops jararaca venom gland. J. Venom. Anim. Toxins Incl. Trop. Dis. 2020, 26, e20190058. [Google Scholar] [CrossRef]
- Serrano, S.M.; Maroun, R.C. Snake venom serine proteinases: Sequence homology vs. substrate specificity, a paradox to be solved. Toxicon 2005, 45, 1115–1132. [Google Scholar] [CrossRef]
- Fox, J.W.; Serrano, S.M. Insights into and speculations about snake venom metalloproteinase (SVMP) synthesis, folding and disulfide bond formation and their contribution to venom complexity. FEBS J. 2008, 275, 3016–3030. [Google Scholar] [CrossRef]
- Matsui, T.; Fujimura, Y.; Titani, K. Snake venom proteases affecting hemostasis and thrombosis. Biochim. Biophys. Acta (BBA)-Protein Struct. Mol. Enzym. 2000, 1477, 146–156. [Google Scholar] [CrossRef]
- Da Silva Cunha, K.C.; Fuly, A.L.; de Araujo, E.G. A phospholipase A(2) isolated from Lachesis muta snake venom increases the survival of retinal ganglion cells in vitro. Toxicon 2011, 57, 580–585. [Google Scholar] [CrossRef]
- Jain, D.; Kumar, S. Snake venom: A potent anticancer agent. Asian Pac. J. Cancer Prev. 2012, 13, 4855–4860. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, N.; Fung, S.Y. Snake Venom L-Amino Acid Oxidases potential biomedical applications. Mal. J. Biochem. Mol. Biol. 2008, 16, 1–10. [Google Scholar]
- Tan, K.K.; Bay, B.H.; Gopalakrishnakone, P. L-amino acid oxidase from snake venom and its anticancer potential. Toxicon 2018, 144, 7–13. [Google Scholar] [CrossRef]
- Pawelek, P.D.; Cheah, J.; Coulombe, R.; Macheroux, P.; Ghisla, S.; Vrielink, A. The structure of L-amino acid oxidase reveals the substrate trajectory into an enantiomerically conserved active site. EMBO J. 2000, 19, 4204–4215. [Google Scholar] [CrossRef] [Green Version]
- Phillips, D.R.; Scarborough, R.M. Clinical Pharmacology of Eptifibatide. Am. J. Cardiol. 1997, 80, 11B–20B. [Google Scholar] [CrossRef]
- Ferreira, S.H. A Bradykinin-Potentiating Factor (Bpf) Present in the Venom of Bothrops Jararca. Br. J. Pharmacol. Chemother. 1965, 24, 163–169. [Google Scholar] [CrossRef] [Green Version]
- Camargo, A.C.; Ianzer, D.; Guerreiro, J.R.; Serrano, S.M. Bradykinin-potentiating peptides: Beyond captopril. Toxicon 2012, 59, 516–523. [Google Scholar] [CrossRef]
- Fox, J.W.; Serrano, S.M. Approaching the golden age of natural product pharmaceuticals from venom libraries: An overview of toxins and toxin-derivatives currently involved in therapeutic or diagnostic applications. Curr. Pharm. Des. 2007, 13, 2927–2934. [Google Scholar] [CrossRef] [Green Version]
- Marsh, N.; Williams, V. Practical applications of snake venom toxins in haemostasis. Toxicon 2005, 45, 1171–1181. [Google Scholar] [CrossRef]
- Vyas, V.K.; Brahmbhatt, K.; Bhatt, H.; Parmar, U. Therapeutic potential of snake venom in cancer therapy: Current perspectives. Asian Pac. J. Trop. Biomed. 2013, 3, 156–162. [Google Scholar] [CrossRef] [Green Version]
- da Silva, R.J.; da Silva, M.G.; Vilela, L.C.; Fecchio, D. Antitumor effect of Bothrops jararaca venom. Mediat. Inflamm. 2002, 11, 99–104. [Google Scholar] [CrossRef]
- Yamazaki, Y.; Morita, T. Snake venom components affecting blood coagulation and the vascular system: Structural similarities and marked diversity. Curr. Pharm. Des. 2007, 13, 2872–2886. [Google Scholar] [CrossRef] [PubMed]
- Dias, M.H.; Kitano, E.S.; Zelanis, A.; Iwai, L.K. Proteomics and drug discovery in cancer. Drug Discov. Today 2016, 21, 264–277. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mora, R.; Valverde, B.; Diaz, C.; Lomonte, B.; Gutierrez, J.M. A Lys49 phospholipase A(2) homologue from Bothrops asper snake venom induces proliferation, apoptosis and necrosis in a lymphoblastoid cell line. Toxicon 2005, 45, 651–660. [Google Scholar] [CrossRef] [PubMed]
- Bateman, E.; Venning, M.; Mirtschin, P.; Woods, A. The effects of selected Australian snake venoms on tumour-associated microvascular endothelial cells (TAMECs) in vitro. J. Venom. Res. 2013, 4, 21–30. [Google Scholar]
- Moreira, V.; Lomonte, B.; Vinolo, M.A.; Curi, R.; Gutierrez, J.M.; Teixeira, C. An Asp49 phospholipase A2 from snake venom induces cyclooxygenase-2 expression and prostaglandin E2 production via activation of NF-kappaB, p38MAPK, and PKC in macrophages. Mediat. Inflamm. 2014, 2014, 105879. [Google Scholar] [CrossRef] [Green Version]
- Leiguez, E.; Giannotti, K.C.; Moreira, V.; Matsubara, M.H.; Gutierrez, J.M.; Lomonte, B.; Rodriguez, J.P.; Balsinde, J.; Teixeira, C. Critical role of TLR2 and MyD88 for functional response of macrophages to a group IIA-secreted phospholipase A2 from snake venom. PLoS ONE 2014, 9, e93741. [Google Scholar] [CrossRef] [Green Version]
- Frejno, M.; Meng, C.; Ruprecht, B.; Oellerich, T.; Scheich, S.; Kleigrewe, K.; Drecoll, E.; Samaras, P.; Hogrebe, A.; Helm, D.; et al. Proteome activity landscapes of tumor cell lines determine drug responses. Nat. Commun. 2020, 11, 3639. [Google Scholar] [CrossRef] [PubMed]
- Ruprecht, B.; Di Bernardo, J.; Wang, Z.; Mo, X.; Ursu, O.; Christopher, M.; Fernandez, R.B.; Zheng, L.; Dill, B.D.; Wang, H.; et al. A mass spectrometry-based proteome map of drug action in lung cancer cell lines. Nat. Chem. Biol. 2020, 16, 1111–1119. [Google Scholar] [CrossRef] [PubMed]
- Furtado, C.M.; Marcondes, M.C.; Sola-Penna, M.; de Souza, M.L.; Zancan, P. Clotrimazole preferentially inhibits human breast cancer cell proliferation, viability and glycolysis. PLoS ONE 2012, 7, e30462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hui, L.; Zheng, Y.; Yan, Y.; Bargonetti, J.; Foster, D.A. Mutant p53 in MDA-MB-231 breast cancer cells is stabilized by elevated phospholipase D activity and contributes to survival signals generated by phospholipase D. Oncogene 2006, 25, 7305–7310. [Google Scholar] [CrossRef] [Green Version]
- Neve, R.M.; Chin, K.; Fridlyand, J.; Yeh, J.; Baehner, F.L.; Fevr, T.; Clark, L.; Bayani, N.; Coppe, J.P.; Tong, F.; et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 2006, 10, 515–527. [Google Scholar] [CrossRef] [Green Version]
- Lv, M.; Shen, Y.; Yang, J.; Li, S.; Wang, B.; Chen, Z.; Li, P.; Liu, P.; Yang, J. Angiomotin Family Members: Oncogenes or Tumor Suppressors? Int. J. Biol. Sci. 2017, 13, 772–781. [Google Scholar] [CrossRef]
- Zhang, H.; Fan, Q. MicroRNA-205 inhibits the proliferation and invasion of breast cancer by regulating AMOT expression. Oncol. Rep. 2015, 34, 2163–2170. [Google Scholar] [CrossRef]
- Couderc, C.; Boin, A.; Fuhrmann, L.; Vincent-Salomon, A.; Mandati, V.; Kieffer, Y.; Mechta-Grigoriou, F.; Del Maestro, L.; Chavrier, P.; Vallerand, D.; et al. AMOTL1 Promotes Breast Cancer Progression and Is Antagonized by Merlin. Neoplasia 2016, 18, 10–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, K.H.; Kim, J.H.; Lee, Y.H.; Bae, H.C.; Lee, H.J.; Woo, S.R.; Oh, S.J.; Lee, K.M.; Yee, C.; Kim, B.W.; et al. Mitochondrial reprogramming via ATP5H loss promotes multimodal cancer therapy resistance. J. Clin. Investig. 2018, 128, 4098–4114. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Z.; Oleinick, N.L.; Zhang, J. ATR/CHK1 inhibitors and cancer therapy. Radiother. Oncol. 2018, 126, 450–464. [Google Scholar] [CrossRef] [PubMed]
- Karnitz, L.M.; Zou, L. Molecular Pathways: Targeting ATR in Cancer Therapy. Clin. Cancer Res. 2015, 21, 4780–4785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Kelly, J.; Chung, A.; Lemp, N.; Chumakova, K.; Yin, D.; Wang, H.J.; Said, J.; Gui, D.; Miller, C.W.; Karlan, B.Y.; et al. Functional domains of CCN1 (Cyr61) regulate breast cancer progression. Int. J. Oncol. 2008, 33, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.T.; Lan, Q.; Lorusso, G.; Duffey, N.; Ruegg, C. The matricellular protein CYR61 promotes breast cancer lung metastasis by facilitating tumor cell extravasation and suppressing anoikis. Oncotarget 2017, 8, 9200–9215. [Google Scholar] [CrossRef] [Green Version]
- Hayes, J.D.; Flanagan, J.U.; Jowsey, I.R. Glutathione transferases. Annu. Rev. Pharmacol. Toxicol. 2005, 45, 51–88. [Google Scholar] [CrossRef] [PubMed]
- Meding, S.; Balluff, B.; Elsner, M.; Schone, C.; Rauser, S.; Nitsche, U.; Maak, M.; Schafer, A.; Hauck, S.M.; Ueffing, M.; et al. Tissue-based proteomics reveals FXYD3, S100A11 and GSTM3 as novel markers for regional lymph node metastasis in colon cancer. J. Pathol. 2012, 228, 459–470. [Google Scholar] [CrossRef] [PubMed]
- Saucedo-Cuevas, L.P.; Ruppen, I.; Ximenez-Embun, P.; Domingo, S.; Gayarre, J.; Munoz, J.; Silva, J.M.; Garcia, M.J.; Benitez, J. CUL4A contributes to the biology of basal-like breast tumors through modulation of cell growth and antitumor immune response. Oncotarget 2014, 5, 2330–2343. [Google Scholar] [CrossRef] [Green Version]
- Lan, F.; Shi, Y. Histone H3.3 and cancer: A potential reader connection. Proc. Natl. Acad. Sci. USA 2015, 112, 6814–6819. [Google Scholar] [CrossRef] [Green Version]
- Ayoubi, H.A.; Mahjoubi, F.; Mirzaei, R. Investigation of the human H3.3B (H3F3B) gene expression as a novel marker in patients with colorectal cancer. J. Gastrointest. Oncol. 2017, 8, 64–69. [Google Scholar] [CrossRef] [Green Version]
- Roberts, E.L.; Newton, R.P.; Axford, A.T. Plasma purine nucleoside phosphorylase in cancer patients. Clin. Chim. Acta 2004, 344, 109–114. [Google Scholar] [CrossRef]
- Vareed, S.K.; Bhat, V.B.; Thompson, C.; Vasu, V.T.; Fermin, D.; Choi, H.; Creighton, C.J.; Gayatri, S.; Lan, L.; Putluri, N.; et al. Metabolites of purine nucleoside phosphorylase (NP) in serum have the potential to delineate pancreatic adenocarcinoma. PLoS ONE 2011, 6, e17177. [Google Scholar] [CrossRef]
- Dummer, R.; Duvic, M.; Scarisbrick, J.; Olsen, E.A.; Rozati, S.; Eggmann, N.; Goldinger, S.M.; Hutchinson, K.; Geskin, L.; Illidge, T.M.; et al. Final results of a multicenter phase II study of the purine nucleoside phosphorylase (PNP) inhibitor forodesine in patients with advanced cutaneous T-cell lymphomas (CTCL) (Mycosis fungoides and Sezary syndrome). Ann. Oncol. 2014, 25, 1807–1812. [Google Scholar] [CrossRef]
- Tan, E.; Besant, P.G.; Zu, X.L.; Turck, C.W.; Bogoyevitch, M.A.; Lim, S.G.; Attwood, P.V.; Yeoh, G.C. Histone H4 histidine kinase displays the expression pattern of a liver oncodevelopmental marker. Carcinogenesis 2004, 25, 2083–2088. [Google Scholar] [CrossRef] [Green Version]
- Besant, P.G.; Attwood, P.V. Histone H4 histidine phosphorylation: Kinases, phosphatases, liver regeneration and cancer. Biochem. Soc. Trans. 2012, 40, 290–293. [Google Scholar] [CrossRef] [Green Version]
- Long, M.; Sun, X.; Shi, W.; Yanru, A.; Leung, S.T.C.; Ding, D.; Cheema, M.S.; MacPherson, N.; Nelson, C.J.; Ausio, J.; et al. A novel histone H4 variant H4G regulates rDNA transcription in breast cancer. Nucleic. Acids Res. 2019, 47, 8399–8409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nacev, B.A.; Feng, L.; Bagert, J.D.; Lemiesz, A.E.; Gao, J.; Soshnev, A.A.; Kundra, R.; Schultz, N.; Muir, T.W.; Allis, C.D. The expanding landscape of ‘oncohistone’ mutations in human cancers. Nature 2019, 567, 473–478. [Google Scholar] [CrossRef] [PubMed]
- Bell, J.L.; Wachter, K.; Muhleck, B.; Pazaitis, N.; Kohn, M.; Lederer, M.; Huttelmaier, S. Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs): Post-transcriptional drivers of cancer progression? Cell. Mol. Life Sci. 2013, 70, 2657–2675. [Google Scholar] [CrossRef] [Green Version]
- Fakhraldeen, S.A.; Clark, R.J.; Roopra, A.; Chin, E.N.; Huang, W.; Castorino, J.; Wisinski, K.B.; Kim, T.; Spiegelman, V.S.; Alexander, C.M. Two Isoforms of the RNA Binding Protein, Coding Region Determinant-binding Protein (CRD-BP/IGF2BP1), Are Expressed in Breast Epithelium and Support Clonogenic Growth of Breast Tumor Cells. J. Biol. Chem. 2015, 290, 13386–13400. [Google Scholar] [CrossRef] [Green Version]
- Kim, T.; Havighurst, T.; Kim, K.; Albertini, M.; Xu, Y.G.; Spiegelman, V.S. Targeting insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) in metastatic melanoma to increase efficacy of BRAF(V600E) inhibitors. Mol. Carcinog. 2018, 57, 678–683. [Google Scholar] [CrossRef] [PubMed]
- Han, W.; Hu, C.; Fan, Z.J.; Shen, G.L. Transcript levels of keratin 1/5/6/14/15/16/17 as potential prognostic indicators in melanoma patients. Sci. Rep. 2021, 11, 1023. [Google Scholar] [CrossRef]
- Palko, E.; Poliska, S.; Sziklai, I.; Penyige, A. Analysis of KRT1, KRT10, KRT19, TP53 and MMP9 expression in pediatric and adult cholesteatoma. PLoS ONE 2018, 13, e0200840. [Google Scholar] [CrossRef]
- Hitzerd, S.M.; Verbrugge, S.E.; Ossenkoppele, G.; Jansen, G.; Peters, G.J. Positioning of aminopeptidase inhibitors in next generation cancer therapy. Amino Acids 2014, 46, 793–808. [Google Scholar] [CrossRef]
- Tian, S.Y.; Chen, S.H.; Shao, B.F.; Cai, H.Y.; Zhou, Y.; Zhou, Y.L.; Xu, A.B. Expression of leucine aminopeptidase 3 (LAP3) correlates with prognosis and malignant development of human hepatocellular carcinoma (HCC). Int. J. Clin. Exp. Pathol. 2014, 7, 3752–3762. [Google Scholar]
- He, X.; Huang, Q.; Qiu, X.; Liu, X.; Sun, G.; Guo, J.; Ding, Z.; Yang, L.; Ban, N.; Tao, T.; et al. LAP3 promotes glioma progression by regulating proliferation, migration and invasion of glioma cells. Int. J. Biol. Macromol. 2015, 72, 1081–1089. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Yang, X.; Shi, H.; Li, M.; Xue, Q.; Ren, H.; Yao, L.; Chen, X.; Zhang, J.; Wang, H. Overexpression of leucine aminopeptidase 3 contributes to malignant development of human esophageal squamous cell carcinoma. J. Mol. Histol. 2014, 45, 283–292. [Google Scholar] [CrossRef]
- Liu, Y.; Yuan, Z.; Song, C. Methylcrotonoyl-CoA carboxylase 2 overexpression predicts an unfavorable prognosis and promotes cell proliferation in breast cancer. Biomark. Med. 2019, 13, 427–436. [Google Scholar] [CrossRef] [PubMed]
- Arco, A.D.; Satrustegui, J. New mitochondrial carriers: An overview. Cell. Mol. Life Sci. 2005, 62, 2204–2227. [Google Scholar] [CrossRef] [PubMed]
- Hao, J.M.; Chen, J.Z.; Sui, H.M.; Si-Ma, X.Q.; Li, G.Q.; Liu, C.; Li, J.L.; Ding, Y.Q.; Li, J.M. A five-gene signature as a potential predictor of metastasis and survival in colorectal cancer. J. Pathol. 2010, 220, 475–489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arigoni, M.; Barutello, G.; Riccardo, F.; Ercole, E.; Cantarella, D.; Orso, F.; Conti, L.; Lanzardo, S.; Taverna, D.; Merighi, I.; et al. miR-135b coordinates progression of ErbB2-driven mammary carcinomas through suppression of MID1 and MTCH2. Am. J. Pathol. 2013, 182, 2058–2070. [Google Scholar] [CrossRef] [Green Version]
- De Palma, G.; Sallustio, F.; Curci, C.; Galleggiante, V.; Rutigliano, M.; Serino, G.; Ditonno, P.; Battaglia, M.; Schena, F.P. The Three-Gene Signature in Urinary Extracellular Vesicles from Patients with Clear Cell Renal Cell Carcinoma. J. Cancer 2016, 7, 1960–1967. [Google Scholar] [CrossRef] [Green Version]
- Nunes, F.D.; de Almeida, F.C.; Tucci, R.; de Sousa, S.C. Homeobox genes: A molecular link between development and cancer. Pesquisa Odontol. Bras. 2003, 17, 94–98. [Google Scholar] [CrossRef] [Green Version]
- Samuel, S.; Naora, H. Homeobox gene expression in cancer: Insights from developmental regulation and deregulation. Eur. J. Cancer 2005, 41, 2428–2437. [Google Scholar] [CrossRef]
- Pyper, S.R.; Viswakarma, N.; Jia, Y.; Zhu, Y.J.; Fondell, J.D.; Reddy, J.K. PRIC295, a Nuclear Receptor Coactivator, Identified from PPARalpha-Interacting Cofactor Complex. PPAR Res. 2010, 2010, 173907. [Google Scholar] [CrossRef] [Green Version]
- Viswakarma, N.; Jia, Y.; Bai, L.; Gao, Q.; Lin, B.; Zhang, X.; Misra, P.; Rana, A.; Jain, S.; Gonzalez, F.J.; et al. The Med1 subunit of the mediator complex induces liver cell proliferation and is phosphorylated by AMP kinase. J. Biol. Chem. 2013, 288, 27898–27911. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, N.; Escalona, R.; Leung, D.; Chan, E.; Kannourakis, G. Tumour microenvironment and metabolic plasticity in cancer and cancer stem cells: Perspectives on metabolic and immune regulatory signatures in chemoresistant ovarian cancer stem cells. Semin. Cancer Biol. 2018, 53, 265–281. [Google Scholar] [CrossRef]
- Levin, A.; Minis, A.; Lalazar, G.; Rodriguez, J.; Steller, H. PSMD5 Inactivation Promotes 26S Proteasome Assembly during Colorectal Tumor Progression. Cancer Res. 2018, 78, 3458–3468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chai, F.L.Y.; Bi, J.; Chen, L.Z.F.; Cui, Y.; Bian, X.; Jiang, J. High expression of REGγ is associated with metastasis and poor prognosis of patients with breast cancer. Int. J. Clin. Exp. Pathol. 2014, 7, 7834. [Google Scholar] [PubMed]
- Chai, F.; Liang, Y.; Bi, J.; Chen, L.; Zhang, F.; Cui, Y.; Jiang, J. REGgamma regulates ERalpha degradation via ubiquitin-proteasome pathway in breast cancer. Biochem. Biophys. Res. Commun. 2015, 456, 534–540. [Google Scholar] [CrossRef]
- Wang, X.; Tu, S.; Tan, J.; Tian, T.; Ran, L.; Rodier, J.F.; Ren, G. REG gamma: A potential marker in breast cancer and effect on cell cycle and proliferation of breast cancer cell. Med. Oncol. 2011, 28, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Coppock, D.; Kopman, C.; Gudas, J.; Cina-Poppe, D.A. Regulation of the quiescence-induced genes: Quiescin Q6, decorin, and ribosomal protein S29. Biochem. Biophys. Res. Commun. 2000, 269, 604–610. [Google Scholar] [CrossRef]
- Notterman, D.A.; Alon, U.; Sierk, A.J.; Levine, A.J. Transcriptional gene expression profiles of colorectal adenoma, adenocarcinoma, and normal tissue examined by oligonucleotide arrays. Cancer Res. 2001, 61, 7. [Google Scholar]
- Lv, Z.; Wang, Z.; Luo, L.; Chen, Y.; Han, G.; Wang, R.; Xiao, H.; Li, X.; Hou, C.; Feng, J.; et al. Spliceosome protein Eftud2 promotes colitis-associated tumorigenesis by modulating inflammatory response of macrophage. Mucosal Immunol. 2019, 12, 1164–1173. [Google Scholar] [CrossRef]
- Sato, N.; Maeda, M.; Sugiyama, M.; Ito, S.; Hyodo, T.; Masuda, A.; Tsunoda, N.; Kokuryo, T.; Hamaguchi, M.; Nagino, M.; et al. Inhibition of SNW1 association with spliceosomal proteins promotes apoptosis in breast cancer cells. Cancer Med. 2015, 4, 268–277. [Google Scholar] [CrossRef] [PubMed]
- Pons, V.; Luyet, P.P.; Morel, E.; Abrami, L.; van der Goot, F.G.; Parton, R.G.; Gruenberg, J. Hrs and SNX3 functions in sorting and membrane invagination within multivesicular bodies. PLoS Biol. 2008, 6, e214. [Google Scholar] [CrossRef]
- Mendelsohn, J. The epidermal growth factor receptor as a target for cancer therapy. Endocr. Relat. Cancer 2001, 8, 3–9. [Google Scholar] [CrossRef] [Green Version]
- John, A.S.; Rothman, V.L.; Tuszynski, G.P. Thrombospondin-1 (TSP-1) Stimulates Expression of Integrin alpha6 in Human Breast Carcinoma Cells: A Downstream Modulator of TSP-1-Induced Cellular Adhesion. J. Oncol. 2010, 2010, 645376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jayachandran, A.; Anaka, M.; Prithviraj, P.; Hudson, C.; McKeown, S.J.; Lo, P.H.; Vella, L.J.; Goding, C.R.; Cebon, J.; Behren, A. Thrombospondin 1 promotes an aggressive phenotype through epithelial-to-mesenchymal transition in human melanoma. Oncotarget 2014, 5, 5782–5797. [Google Scholar] [CrossRef] [Green Version]
- Tzeng, H.T.; Tsai, C.H.; Yen, Y.T.; Cheng, H.C.; Chen, Y.C.; Pu, S.W.; Wang, Y.S.; Shan, Y.S.; Tseng, Y.L.; Su, W.C.; et al. Dysregulation of Rab37-Mediated Cross-talk between Cancer Cells and Endothelial Cells via Thrombospondin-1 Promotes Tumor Neovasculature and Metastasis. Clin. Cancer Res. 2017, 23, 2335–2345. [Google Scholar] [CrossRef] [Green Version]
- Shi, H.; Hayes, M.; Kirana, C.; Miller, R.; Keating, J.; Macartney-Coxson, D.; Stubbs, R. TUFM is a potential new prognostic indicator for colorectal carcinoma. Pathology 2012, 44, 506–512. [Google Scholar] [CrossRef]
- Weng, X.; Zheng, S.; Shui, H.; Lin, G.; Zhou, Y. TUFM-knockdown inhibits the migration and proliferation of gastrointestinal stromal tumor cells. Oncol. Lett. 2020, 20, 250. [Google Scholar] [CrossRef]
- He, K.; Guo, X.; Liu, Y.; Li, J.; Hu, Y.; Wang, D.; Song, J. TUFM downregulation induces epithelial-mesenchymal transition and invasion in lung cancer cells via a mechanism involving AMPK-GSK3beta signaling. Cell. Mol. Life Sci. 2016, 73, 2105–2121. [Google Scholar] [CrossRef] [PubMed]
- Kulawiec, M.; Arnouk, H.; Desouki, M.M.; Kazim, L.; Still, I.; Singh, K.K. Proteomic analysis of mitochondria-to-nucleus retrograde response in human cancer. Cancer Biol. Ther. 2006, 5, 967–975. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wubulikasimu, G.; Zhao, X.; Wang, C.; Liu, R.; Wang, L.; Zhu, X.; Chen, Z. UQCRC1 downregulation is correlated with lymph node metastasis and poor prognosis in CRC. Eur. J. Surg. Oncol. 2019, 45, 1005–1010. [Google Scholar] [CrossRef]
- Wang, Q.; Li, M.; Gan, Y.; Jiang, S.; Qiao, J.; Zhang, W.; Fan, Y.; Shen, Y.; Song, Y.; Meng, Z.; et al. Mitochondrial Protein UQCRC1 is Oncogenic and a Potential Therapeutic Target for Pancreatic Cancer. Theranostics 2020, 10, 2141–2157. [Google Scholar] [CrossRef] [PubMed]
- Mazure, N.M. VDAC in cancer. Biochim. Biophys. Acta Bioenerg. 2017, 1858, 665–673. [Google Scholar] [CrossRef]
- Sotgia, F.; Fiorillo, M.; Lisanti, M.P. Mitochondrial markers predict recurrence, metastasis and tamoxifen-resistance in breast cancer patients: Early detection of treatment failure with companion diagnostics. Oncotarget 2017, 8, 68730–68745. [Google Scholar] [CrossRef] [Green Version]
- Camara, A.K.S.; Zhou, Y.; Wen, P.C.; Tajkhorshid, E.; Kwok, W.M. Mitochondrial VDAC1: A Key Gatekeeper as Potential Therapeutic Target. Front. Physiol. 2017, 8, 460. [Google Scholar] [CrossRef] [Green Version]
- Magri, A.; Reina, S.; De Pinto, V. VDAC1 as Pharmacological Target in Cancer and Neurodegeneration: Focus on Its Role in Apoptosis. Front. Chem. 2018, 6, 108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thul, P.J.; Akesson, L.; Wiking, M.; Mahdessian, D.; Geladaki, A.; Ait Blal, H.; Alm, T.; Asplund, A.; Bjork, L.; Breckels, L.M.; et al. A subcellular map of the human proteome. Science 2017, 356. [Google Scholar] [CrossRef]
- Uhlen, M.; Fagerberg, L.; Hallstrom, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, A.; Kampf, C.; Sjostedt, E.; Asplund, A.; et al. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef] [PubMed]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjostedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017, 357. [Google Scholar] [CrossRef] [Green Version]
- Doncheva, N.T.; Morris, J.H.; Gorodkin, J.; Jensen, L.J. Cytoscape StringApp: Network Analysis and Visualization of Proteomics Data. J. Proteome Res. 2019, 18, 623–632. [Google Scholar] [CrossRef]
- Essex, H.E.; Priestley, J.T. Effect of Rattlesnake Venom on Flexner-Jobling’s Carcinoma in the White Rat (Mus Norvegicus Albinus.). Exp. Biol. Med. 1931, 28, 550–551. [Google Scholar] [CrossRef]
- Ferreira, S.H.; Greene, L.H.; Alabaster, V.A.; Bakhle, Y.S.; Vane, J.R. Activity of various fractions of bradykinin potentiating factor against angiotensin I converting enzyme. Nature 1970, 225, 379–380. [Google Scholar] [CrossRef] [PubMed]
- Cushman, D.W.; Ondetti, M.A. History of the design of captopril and related inhibitors of angiotensin converting enzyme. Hypertension 1991, 17, 589–592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, C.G.; Vane, J.R. The discovery of captopril. FASEB J. 2003, 17, 788–789. [Google Scholar] [CrossRef] [PubMed]
- Ma, R.; Mahadevappa, R.; Kwok, H.F. Venom-based peptide therapy: Insights into anti-cancer mechanism. Oncotarget 2017, 8, 100908–100930. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Huang, J.; Lin, Y. Snake Venoms in Cancer Therapy: Past, Present and Future. Toxins 2018, 10, 346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sukocheva, O.; Menschikowski, M.; Hagelgans, A.; Yarla, N.S.; Siegert, G.; Reddanna, P.; Bishayee, A. Current insights into functions of phospholipase A2 receptor in normal and cancer cells: More questions than answers. Semin. Cancer Biol. 2019, 56, 116–127. [Google Scholar] [CrossRef]
- Peng, Z.; Chang, Y.; Fan, J.; Ji, W.; Su, C. Phospholipase A2 superfamily in cancer. Cancer Lett. 2021, 497, 165–177. [Google Scholar] [CrossRef]
- Cathcart, J.; Pulkoski-Gross, A.; Cao, J. Targeting Matrix Metalloproteinases in Cancer: Bringing New Life to Old Ideas. Genes Dis. 2015, 2, 26–34. [Google Scholar] [CrossRef] [Green Version]
- Di Cara, G.; Marabeti, M.R.; Musso, R.; Riili, I.; Cancemi, P.; Pucci Minafra, I. New Insights into the Occurrence of Matrix Metalloproteases-2 and -9 in a Cohort of Breast Cancer Patients and Proteomic Correlations. Cells 2018, 7, 89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quintero-Fabian, S.; Arreola, R.; Becerril-Villanueva, E.; Torres-Romero, J.C.; Arana-Argaez, V.; Lara-Riegos, J.; Ramirez-Camacho, M.A.; Alvarez-Sanchez, M.E. Role of Matrix Metalloproteinases in Angiogenesis and Cancer. Front. Oncol. 2019, 9, 1370. [Google Scholar] [CrossRef] [Green Version]
- Bezerra, P.H.A.; Ferreira, I.M.; Franceschi, B.T.; Bianchini, F.; Ambrosio, L.; Cintra, A.C.O.; Sampaio, S.V.; de Castro, F.A.; Torqueti, M.R. BthTX-I from Bothrops jararacussu induces apoptosis in human breast cancer cell lines and decreases cancer stem cell subpopulation. J. Venom. Anim. Toxins Incl. Trop. Dis. 2019, 25, e20190010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murray, A.S.; Hyland, T.E.; Sala-Hamrick, K.E.; Mackinder, J.R.; Martin, C.E.; Tanabe, L.M.; Varela, F.A.; List, K. The cell-surface anchored serine protease TMPRSS13 promotes breast cancer progression and resistance to chemotherapy. Oncogene 2020, 39, 6421–6436. [Google Scholar] [CrossRef] [PubMed]
- Krasny, L.; Huang, P.H. Data-independent acquisition mass spectrometry (DIA-MS) for proteomic applications in oncology. Mol. Omics 2021, 17, 29–42. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Li, J.; Chen, M.M.; Luo, Y.; Ju, Z.; Nesser, N.K.; Johnson-Camacho, K.; Boniface, C.T.; Lawrence, Y.; Pande, N.T.; et al. Large-Scale Characterization of Drug Responses of Clinically Relevant Proteins in Cancer Cell Lines. Cancer Cell 2020, 38, 829–843.e4. [Google Scholar] [CrossRef]
- Klaeger, S.; Heinzlmeir, S.; Wilhelm, M.; Polzer, H.; Vick, B.; Koenig, P.A.; Reinecke, M.; Ruprecht, B.; Petzoldt, S.; Meng, C.; et al. The target landscape of clinical kinase drugs. Science 2017, 358. [Google Scholar] [CrossRef] [Green Version]
- Qi, Z.; Zhang, L.; Chen, Y.; Ma, X.; Gao, X.; Du, J.; Zhang, F.; Cheng, X.; Cui, W. Biological variations of seven tumor markers. Clin. Chim. Acta 2015, 450, 233–236. [Google Scholar] [CrossRef]
- Lamond, A.I.; Mann, M. Cell biology and the genome projects a concerted strategy for characterizing multiprotein complexes by using mass spectrometry. Trends Cell Biol. 1997, 7, 139–142. [Google Scholar] [CrossRef]
- Tavares, C.; Maciel, T.; Burin, S.; Ambrosio, L.; Ghisla, S.; Sampaio, S.; Castro, F. l-Amino acid oxidase isolated from Calloselasma rhodostoma snake venom induces cytotoxicity and apoptosis in JAK2V617F-positive cell lines. Rev. Bras. Hematol. Hemoter. 2016, 38, 128–134. [Google Scholar] [CrossRef] [Green Version]
- Zakraoui, O.; Marcinkiewicz, C.; Aloui, Z.; Othman, H.; Grepin, R.; Haoues, M.; Essafi, M.; Srairi-Abid, N.; Gasmi, A.; Karoui, H.; et al. Lebein, a snake venom disintegrin, suppresses human colon cancer cells proliferation and tumor-induced angiogenesis through cell cycle arrest, apoptosis induction and inhibition of VEGF expression. Mol. Carcinog. 2017, 56, 18–35. [Google Scholar] [CrossRef] [PubMed]
- Bernardes-Oliveira, E.; Gomes, D.L.; Martelli Palomino, G.; Juvenal Silva Farias, K.; da Silva, W.D.; Rocha, H.A.; Goncalves, A.K.; Fernandes-Pedrosa, M.F.; Crispim, J.C. Bothrops jararaca and Bothrops erythromelas Snake Venoms Promote Cell Cycle Arrest and Induce Apoptosis via the Mitochondrial Depolarization of Cervical Cancer Cells. Evid. Based Complement. Altern. Med. 2016, 2016, 1574971. [Google Scholar] [CrossRef]
- Knorre, D.G.; Kudryashova, N.V.; Godovikova, T.S. Chemical and functional aspects of posttranslational modification of proteins. Acta Nat. 2009, 1, 23. [Google Scholar]
- Freed, E.F.; Bleichert, F.; Dutca, L.M.; Baserga, S.J. When ribosomes go bad: Diseases of ribosome biogenesis. Mol. Biosyst 2010, 6, 481–493. [Google Scholar] [CrossRef]
- Ruggero, D.; Pandolfi, P.P. Does the ribosome translate cancer? Nat. Rev. Cancer 2003, 3, 179–192. [Google Scholar] [CrossRef]
- Bilanges, B.; Stokoe, D. Mechanisms of translational deregulation in human tumors and therapeutic intervention strategies. Oncogene 2007, 26, 5973–5990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hershko, A. Ubiquitin: Roles in protein modification and breakdown. Cell 1983, 34, 11–12. [Google Scholar] [CrossRef]
- Hershko, A.; Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 1998, 67, 425–479. [Google Scholar] [CrossRef]
- Jankowska, E.; Stoj, J.; Karpowicz, P.; Osmulski, P.A.; Gaczynska, M. The proteasome in health and disease. Curr. Pharm. Des. 2013, 19, 19. [Google Scholar]
- Pajonk, F.; McBride, W.H. The Proteasome in Cancer Biology and Treatment. Radiat. Res. 2001, 156, 447–459. [Google Scholar] [CrossRef]
- Teicher, B.A.; Ara, G.; Herbst, R.; Palombella, V.J.; Adams, J. The proteasome inhibitor PS-341 in cancer therapy. Clin. Cancer Res. 1999, 5, 2638–2645. [Google Scholar] [PubMed]
- Thul, P.J.; Lindskog, C. The human protein atlas: A spatial map of the human proteome. Protein Sci. 2018, 27, 233–244. [Google Scholar] [CrossRef] [Green Version]
- Dias, M.H.; Fonseca, C.S.; Zeidler, J.D.; Albuquerque, L.L.; da Silva, M.S.; Cararo-Lopes, E.; Reis, M.S.; Noel, V.; Dos Santos, E.O.; Prior, I.A.; et al. Fibroblast Growth Factor 2 lethally sensitizes cancer cells to stress-targeted therapeutic inhibitors. Mol. Oncol. 2019, 13, 290–306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montoni, F.; Andreotti, D.Z.; Eichler, R.; Santos, W.D.S.; Kisaki, C.Y.; Arcos, S.S.S.; Lima, I.F.; Soares, M.A.M.; Nishiyama-Jr, M.Y.; Nava-Rodrigues, D.; et al. The impact of rattlesnake venom on mice cerebellum proteomics points to synaptic inhibition and tissue damage. J. Proteom. 2020, 221, 103779. [Google Scholar] [CrossRef]
- Mi, H.; Muruganujan, A.; Huang, X.; Ebert, D.; Mills, C.; Guo, X.; Thomas, P.D. Protocol Update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0). Nat. Protoc. 2019, 14, 703–721. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Poudel, S.; Muruganujan, A.; Casagrande, J.T.; Thomas, P.D. PANTHER version 10: Expanded protein families and functions, and analysis tools. Nucleic Acids Res. 2016, 44, D336–D342. [Google Scholar] [CrossRef] [Green Version]
- Thomas, P.D.; Campbell, M.J.; Kejariwal, A.; Mi, H.; Karlak, B.; Daverman, R.; Diemer, K.; Muruganujan, A.; Narechania, A. PANTHER: A library of protein families and subfamilies indexed by function. Genome Res. 2003, 13, 2129–2141. [Google Scholar] [CrossRef] [Green Version]
- Thomas, P.D.; Kejariwal, A.; Campbell, M.J.; Mi, H.; Diemer, K.; Guo, N.; Ladunga, I.; Ulitsky-Lazareva, B.; Muruganujan, A.; Rabkin, S.; et al. PANTHER: A browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic Acids Res. 2003, 31, 334–341. [Google Scholar] [CrossRef]
- Cox, J.; Hein, M.Y.; Luber, C.A.; Paron, I.; Nagaraj, N.; Mann, M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell Proteom. 2014, 13, 2513–2526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef]
- Lê, S.; Josse, J.; Husson, F. FactoMineR: AnRPackage for Multivariate Analysis. J. Stat. Softw. 2008, 25. [Google Scholar] [CrossRef] [Green Version]






| Protein | Protein Name | Protein Description Related to Cancer | Cancer Type Association | References |
|---|---|---|---|---|
| AMOT | Angiomotin | Plays a central role in tight junction maintenance. Appears to regulate endothelial cell migration and tube formation. May also play a role in the assembly of endothelial cell-cell junctions. Plays a critical role in angiogenesis, proliferation and migration and invasion of cancer cells | BL, BR, CE, CL, CR, EN, HN, KD, LE, LI, LA, LS, OV, PR, ST | [49,50,51] |
| ATP5PD | ATP synthase peripheral stalk subunit D | Mitochondrial ATP synthase catalyzes ATP synthesis, utilizing an electrochemical gradient of protons across the inner membrane during oxidative phosphorylation. Linked to failure of therapy, disease progression, and poor survival in patients with cancer. High expression of ATP5PD has been observed in several types of cancer | BR, CE, CL, EN, GL, HN, LI, LU, LY, ME, OV, PA, PR, RE, SK, TE, TY | [52] |
| ATR | Serine/threonine-protein kinase | Plays important roles for cell survival and is considered a major mediator of DNA response in human cells, preventing cells with damaged or incompletely replicated DNA from entering mitosis when cells are damaged by radiotherapy or chemotherapy during cancer treatment | BL, BR, CE, CR, EN, GB, HN, KD, LU, LA, LS, OV, ST, TY | [53,54] |
| CYR61 | Cysteine-rich heparin-binding protein 61 | Plays an important role in cell proliferation, survival, chemotaxis, angiogenesis, adhesion, and migration of different types of cells. Participate in key different cellular events during vascular development, angiogenesis, wound healing and the development and progression of various types of cancers | BN, BR, CR, EN, GA, GB, GL, LI, LU, OV, PA, PR, ST, TE, UR | [55,56] |
| GSTM3 | The glutathione S-transferase Mu 3 | Part of the GSTs enzymes that have functions such as immunological system evasion and inhibition of apoptosis. Involved in prostaglandin and leukotriene synthesis and metabolization of both endogenous compounds and xenobiotics such as chemotherapeutic drugs, insecticides, carcinogens, and oxidative stress byproducts | BL, BR, CR, EN, LE, LA, LS, OV, PA, ST, TY, UR | [57,58] |
| H3F3B/H3C15 | H3 histone family member 3B | Core component of nucleosome. Histones play a central role in transcription regulation, DNA repair, DNA replication and chromosomal stability and are related to different types of cancer. H3F3B mutation has been described to lead to some human cancers | BL, BN, BR, CE, CH, CR, EN, GB, HN, LU, OV, UR | [59,60,61] |
| HEL-S-156an PNP | Purine nucleoside phosphorylase | Catalyze the phosphorolysis of purine nucleosides. Mutations which result in nucleoside phosphorylase deficiency result in defective T-cell (cell-mediated) immunity but can also affect B-cell immunity and antibody responses. High expression of PNP has been observed in several types of cancer | BR, CL, CR, GA, GL, KD, LI, LU, LA, LY, ME, OV, PR, TY | [62,63,64] |
| HIST1H4J | Histone H4 | Core component of nucleosome. Histones play a central role in transcription regulation, DNA repair, DNA replication and chromosomal stability. Post-translational alterations of histones have been shown to affect the activation and repression of oncogenes and tumor suppressor genes | BR, CE, CR, GL, HN, LI, LU, ME, OV, PA, PR, SK, ST, TE, TY | [65,66,67,68] |
| IGF2BP1 | Insulin like growth factor 2 MRNA binding protein 1 | RNA-binding factor that recruits target transcripts to cytoplasmic protein–RNA complexes (mRNPs). IGF2BP1 has an oncogenic role, characterized by changes in actin dynamics, migration, invasion, proliferation, and self-renewal. Play a role in resistance to drugs | BR, CE, CR, EN, GB, HN, LI, LU, LA, LS, ME, OV, PR, ST, TE, UR | [69,70,71] |
| KRT1 | Keratin | May regulate the activity of kinases such as PKC and SRC via binding to integrin beta-1 (ITB1) and to the receptor of activated protein C kinase 1 (RACK1). High expression of KRT1 protein has been observed in several types of cancer and is correlated with advanced melanoma tumor stage and infiltration of immune cells | BR, CE, CR, EN, GB, HN, KD, LI, LS, LA, OV, SK, ST, UR | [72,73] |
| LAP3 | Leucine aminopeptidase 3 | Cytosolic metallopeptidase that catalyzes the removal of unsubstituted N-terminal hydrophobic amino acids from various peptides. Involved in the metabolism of glutathione and in the degradation of glutathione S-conjugates, which may play a role in the control of the cell redox status. Related to protein renewal. Have a potential for determining the prognosis for breast cancer | BL, BR, CR, EN, HN, KD, LI, LA, LS, PA, ST, TY | [74,75,76,77] |
| MCCC2 | Methylcrotonoyl-CoA carboxylase beta chain, mitochondrial | Enzyme that catalyzes the conversion of 3-methylcrotonyl-CoA to 3-methylglutaconyl-CoA, a critical step for leucine and isovaleric acid catabolism. Overexpression of MCCC2 is associated with tumor stage, node, metastasis, lymph node metastasis and predicts unfavorable prognosis. Additionally, involved in the development and formation of some tumors, such as breast cancer | BL, BR, CR, EN, HN, KD, LI, LA, OV, PA, PR, ST, TY | [78] |
| MTCH2 | Mitochondrial carrier homolog 2 | Member of the SLC25 family of nuclear-encoded transporters that are localized in the inner mitochondrial membrane. Members of this superfamily are involved in many metabolic pathways and cell functions. Associated with metastasis and tumor cell survival. Indirect involvement in the expression of miR-135b mRNA, which is one of the proteins responsible for tumorigenicity | BL, BR, CR, EN, HN, KD, LI, LA, ME, OV, PA, PR, ST, TY | [79,80,81] |
| PCBD | 4a-hydroxytetrahydrobiopterin dehydratase | Involved in tetrahydrobiopterin biosynthesis. Regulates various aspects of cell morphogenesis and differentiation as a cofactor for the homeobox transcription factor. Several types of cancer show expression or alteration in the homeobox genes. PCBD degradation increases cell survival and proliferation, and inhibits tumor cell differentiation | BR, CL, CR, EN, LE, LI, LU, OV, PA, PR, RE, SK | [82,83,84] |
| PRIC295 | Peroxisome proliferator-activated receptor-a (PPARα)-interacting cofactor | Functions as a transcriptional coactivator for nuclear receptors. Enhances the activation of PPARα and PPARγ and plays a key role in lipid metabolism and energy combustion regulating the genes for fatty acid oxidation. Observed to be significantly enhanced in chemotherapy recurrence when compared to chemotherapy treatment in ovarian cancer patients | BL, BR, CR, EN, GB, HN, KD, LE, LA, LS, ME, OV, PA, PR, ST, TY | [85,86,87] |
| PSMD5 | The 26S proteasome non-ATPase regulatory subunit 5 | Acts as a chaperone during the assembly of the 26S proteasome. Expression reduced in several types of cancer including intestinal and colorectal tumors | BL, BR, CR, EN, HN, PR, ST, TY | [88] |
| PSME2 | The proteasome activator complex subunit 2 | Implicated in immunoproteasome assembly and required for efficient antigen processing. Member of the PSME family that regulates proteasome function. Elevated expression of PSME have also been associated with several types of cancer | BR, CR, EN, HN, LC, LA, LS, ME, PR, ST | [89,90,91] |
| RPS29 | Ribosomal protein S29 | Belongs to the universal ribosomal protein uS14 family. Related to have tumor suppressor activity for ras-transformed NIH3T3 cells. High expression RPS29 mRNA levels observed in adenomas | BR, CE, CR, EN, HN, KD, LA, OV, ST | [92,93] |
| SNRP116/EFTUD2 | Small nuclear ribonucleoprotein component | Required for pre-mRNA splicing as a component of the spliceosome, including pre-catalytic, catalytic, and post-catalytic spliceosomal complexes. Knockout of EFTUD2 suppressed the development and tumor progression due to impaired activation of NF-kB signaling in macrophages | BL, BR, CR, EN, GB, HN, KD, LA, LS, ME, OV, PR, ST, TE, TY, UR | [94,95] |
| SNX3 | The sorting nexin 3 | Phosphoinositide-binding protein required for multivesicular body formation. Plays a role in protein transport between cellular compartments. The knockdown of SNX3 is associated with degradation of the EGF receptor which is related to resistance to chemotherapy and radiotherapy | BR, CR, EN, GL, HN, KD, LU, OV, RE, TY | [96,97] |
| THBS1 | Thrombospondin 1 | Adhesive glycoprotein that mediates cell-to-cell and cell-to-matrix interactions. Influences angiogenesis modulation by regulating adhesion, invasion, metastasis, migration, proliferation, and apoptosis and has been implicated in numerous types of cancers | BR, CR, EN, ES, HN, KD, LA, LS, LY, ME, OV, PA, PR, SK, ST, TE, TY | [98,99,100] |
| TUFM | Tu translation elongation factor, mitochondrial | Promotes the GTP-dependent binding of aminoacyl-tRNA to the A-site of ribosomes during protein biosynthesis. Plays important roles in the regulation of autophagy and innate immunity. TUFM is highly expressed in several types of cancers | BR, CR, EN, ES, GA, GS, HN, LI, LU, LA, ME, PA, PR, RE, SK, ST, TE | [101,102,103] |
| UQCRC1 | The ubiquinol-cytochrome C reductase core protein 1 | Component of the ubiquinol-cytochrome c reductase complex, which is part of the mitochondrial respiratory chain. High expression was observed in several types of cancer. Negative expression correlated significantly with clinical and pathological parameters including tumor stage, vascular invasion, and lymph node metastasis, suggesting that the reduction of this protein is associated with tumor progression | BR, CR, EN, GB, GL, HN, LI, LA, LY, ME, OV, PA, PR, RE, ST, TE, TY, UR | [104,105,106] |
| VDAC1 and VDAC2 | The voltage-dependent anion selective channel 1 and 2 | Forms a channel through the mitochondrial outer membrane and plasma membrane allowing diffusion of small hydrophilic molecules. In the plasma membrane it is involved in cell volume regulation and apoptosis. The abnormal expression or mal functioning of VDACs has been reported in multiple tumors and it has been considered as a biomarker capable of predicting treatment failure and breast cancer recurrence | BL, BR, CR, EN, GB, HN, LI, LA, LS, ME, OV, PA, PR, RE, ST, TY, UR | [107,108,109,110] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kisaki, C.Y.; Arcos, S.S.S.; Montoni, F.; da Silva Santos, W.; Calacina, H.M.; Lima, I.F.; Cajado-Carvalho, D.; Ferro, E.S.; Nishiyama-Jr, M.Y.; Iwai, L.K. Bothrops Jararaca Snake Venom Modulates Key Cancer-Related Proteins in Breast Tumor Cell Lines. Toxins 2021, 13, 519. https://doi.org/10.3390/toxins13080519
Kisaki CY, Arcos SSS, Montoni F, da Silva Santos W, Calacina HM, Lima IF, Cajado-Carvalho D, Ferro ES, Nishiyama-Jr MY, Iwai LK. Bothrops Jararaca Snake Venom Modulates Key Cancer-Related Proteins in Breast Tumor Cell Lines. Toxins. 2021; 13(8):519. https://doi.org/10.3390/toxins13080519
Chicago/Turabian StyleKisaki, Carolina Yukiko, Stephanie Santos Suehiro Arcos, Fabio Montoni, Wellington da Silva Santos, Hamida Macêdo Calacina, Ismael Feitosa Lima, Daniela Cajado-Carvalho, Emer Suavinho Ferro, Milton Yutaka Nishiyama-Jr, and Leo Kei Iwai. 2021. "Bothrops Jararaca Snake Venom Modulates Key Cancer-Related Proteins in Breast Tumor Cell Lines" Toxins 13, no. 8: 519. https://doi.org/10.3390/toxins13080519