CD44-Targeted Lipid Polymer Hybrid Nanoparticles Enhance Anti-Breast Cancer Effect of Cordyceps militaris Extracts
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Cultivation and Extraction of CM Fruiting Body
2.3. Analysis of CM Extract (CME)
2.4. Modification of PGA
2.5. Characterization of Polymers
2.6. Preparation of LPNPs
2.7. Characterization of LPNPs
2.8. Release Study
2.9. Cytotoxicity Study
2.10. Cellular Uptake Study
2.11. Statistical Analysis
3. Results and Discussion
3.1. Extraction of CM Fruiting Bodies
3.2. Modification of PGA Polymers
3.3. Characteristics of CME-Loaded LPNPs with and without HYA Conjugation
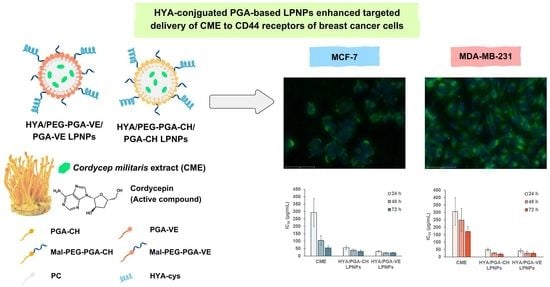
3.4. Cytotoxicity of CME-Loaded LPNPs
3.5. Cellular Uptake
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Das, S.K.; Masuda, M.; Sakurai, A.; Sakakibara, M. Medicinal uses of the mushroom Cordyceps militaris: Current state and prospects. Fitoterapia 2010, 81, 961–968. [Google Scholar] [CrossRef]
- Olatunji, O.J.; Tang, J.; Tola, A.; Auberon, F.; Oluwaniyi, O.; Ouyang, Z. The genus Cordyceps: An extensive review of its traditional uses, phytochemistry and pharmacology. Fitoterapia 2018, 129, 293–316. [Google Scholar] [CrossRef]
- Tuli, H.S.; Sandhu, S.S.; Sharma, A.K. Pharmacological and therapeutic potential of Cordyceps with special reference to cordycepin. 3 Biotech 2014, 4, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Kondrashov, A.; Meijer, H.A.; Barthet-Barateig, A.; Parker, H.N.; Khurshid, A.; Tessier, S.; Sicard, M.; Knox, A.J.; Pang, L.; De Moor, C.H. Inhibition of polyadenylation reduces inflammatory gene induction. RNA 2012, 18, 2236–2250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, M.L.; Park, S.Y.; Kim, Y.H.; Oh, J.I.; Lee, S.J.; Park, G. The neuroprotective effects of cordycepin inhibit glutamate-induced oxidative and ER stress-associated apoptosis in hippocampal HT22 cells. Neurotoxicology 2014, 41, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.Y.; Park, S.J.; Park, Y.J. Anticancer effects of Cordyceps militaris extract in human ovarian cancer cells via ADORA2B (P05-012-19). Curr. Dev. Nutr. 2019, 3, nzz030-P05. [Google Scholar] [CrossRef] [Green Version]
- Lee, D.; Lee, W.-Y.; Jung, K.; Kwon, Y.S.; Kim, D.; Hwang, G.S.; Kim, C.-E.; Lee, S.; Kang, K.S. The inhibitory effect of cordycepin on the proliferation of MCF-7 breast cancer cells, and its mechanism: An investigation using network pharmacology-based analysis. Biomolecules 2019, 9, 414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bizarro, A.; Ferreira, I.C.; Soković, M.; van Griensven, L.J.; Sousa, D.; Vasconcelos, M.H.; Lima, R.T. Cordyceps militaris (L.) link fruiting body reduces the growth of a non-small cell lung cancer cell line by increasing cellular levels of p53 and p21. Molecules 2015, 20, 13927–13940. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.H.; Lee, S.; Lee, K.; Shin, Y.S.; Kang, H.; Cho, H. Anti-cancer effect of Cordyceps militaris in human colorectal carcinoma RKO cells via cell cycle arrest and mitochondrial apoptosis. Daru 2015, 23, 35. [Google Scholar] [CrossRef] [Green Version]
- Ruma, I.M.W.; Putranto, E.W.; Kondo, E.; Watanabe, R.; Saito, K.; Inoue, Y.; Yamamoto, K.-I.; Nakata, S.; Kaihata, M.; Murata, H.; et al. Extract of Cordyceps militaris inhibits angiogenesis and suppresses tumor growth of human malignant melanoma cells. Int. J. Oncol. 2014, 45, 209–218. [Google Scholar] [CrossRef] [Green Version]
- Jo, E.; Jang, H.-J.; Yang, K.E.; Jang, M.S.; Huh, Y.H.; Yoo, H.-S.; Park, J.; Jang, I.-S.; Park, S.J. Cordyceps militaris exerts antitumor effect on carboplatin-resistant ovarian cancer via activation of ATF3/TP53 signaling in vitro and in vivo. Nat. Prod. Commun. 2020, 15, 1934578X20902558. [Google Scholar] [CrossRef] [Green Version]
- Tsai, Y.-J.; Lin, L.-C.; Tsai, T.-H. Pharmacokinetics of Adenosine and Cordycepin, a Bioactive Constituent of Cordyceps sinensis in Rat. J. Agric. Food Chem. 2010, 58, 4638–4643. [Google Scholar] [CrossRef]
- Rodman, L.E.; Farnell, D.R.; Coyne, J.M.; Allan, P.W.; Hill, D.L.; Duncan, K.L.; Tomaszewski, J.E.; Smith, A.C.; Page, J.G. Toxicity of cordycepin in combination with the adenosine deaminase inhibitor 2′-deoxycoformycin in beagle dogs. Toxicol. Appl. Pharmacol. 1997, 147, 39–45. [Google Scholar] [CrossRef]
- Babu, A.; Templeton, A.; Munshi, A.; Ramesh, R. Nanodrug Delivery Systems: A Promising Technology for Detection, Diagnosis, and Treatment of Cancer. AAPS Pharm. Sci. Tech. 2014, 15, 709–721. [Google Scholar] [CrossRef] [Green Version]
- Damrongrak, K.; Kloysawat, K.; Bunsupa, S.; Sakchasri, K.; Wongrakpanich, A.; Taresco, V.; Cuzzucoli Crucitti, V.; Garnett, M.C.; Suksiriworapong, J. Delivery of acetogenin-enriched Annona muricata Linn leaf extract by folic acid-conjugated and triphenylphosphonium-conjugated poly(glycerol adipate) nanoparticles to enhance toxicity against ovarian cancer cells. Int. J. Pharm. 2022, 618, 121636. [Google Scholar] [CrossRef]
- Suksiriworapong, J.; Phoca, K.; Ngamsom, S.; Sripha, K.; Moongkarndi, P.; Junyaprasert, V.B. Comparison of poly(ε-caprolactone) chain lengths of poly(ε-caprolactone)-co-d-α-tocopheryl-poly(ethylene glycol) 1000 succinate nanoparticles for enhancement of quercetin delivery to SKBR3 breast cancer cells. Eur. J. Pharm. Biopharm. 2016, 101, 15–24. [Google Scholar] [CrossRef]
- Brambilla, D.; Luciani, P.; Leroux, J.-C. Breakthrough discoveries in drug delivery technologies: The next 30years. J. Control. Release 2014, 190, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Maeda, H.; Nakamura, H.; Fang, J. The EPR effect for macromolecular drug delivery to solid tumors: Improvement of tumor uptake, lowering of systemic toxicity, and distinct tumor imaging in vivo. Adv. Drug Del. Rev. 2013, 65, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Ballesta, M.; Gil-Izquierdo, Á.; García-Viguera, C.; Domínguez-Perles, R. Nanoparticles and Controlled Delivery for Bioactive Compounds: Outlining Challenges for New “Smart-Foods” for Health. Foods 2018, 7, 72. [Google Scholar] [CrossRef] [Green Version]
- Chavda, V.P.; Patel, A.B.; Mistry, K.J.; Suthar, S.F.; Wu, Z.-X.; Chen, Z.-S.; Hou, K. Nano-Drug Delivery Systems Entrapping Natural Bioactive Compounds for Cancer: Recent Progress and Future Challenges. Front. Oncol. 2022, 12, 867655. [Google Scholar] [CrossRef] [PubMed]
- Mattheolabakis, G.; Milane, L.; Singh, A.; Amiji, M.M. Hyaluronic acid targeting of CD44 for cancer therapy: From receptor biology to nanomedicine. J. Drug Target. 2015, 23, 605–618. [Google Scholar] [CrossRef]
- Thummarati, P.; Suksiriworapong, J.; Sakchaisri, K.; Junyaprasert, V.B. Effect of chemical linkers of curcumin conjugated hyaluronic acid on nanoparticle properties and in vitro performances in various cancer cells. J. Drug Deliv. Sci. Technol. 2021, 61, 102323. [Google Scholar] [CrossRef]
- Lin, W.J.; Lee, W.-C.; Shieh, M.-J. Hyaluronic acid conjugated micelles possessing CD44 targeting potential for gene delivery. Carbohydr. Polym. 2017, 155, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Wang, Y.; Wang, L.; Liang, Z.; Li, D.; Xu, X.; Chen, Y.; Yang, X.; Zhang, H.; Niu, H. Self-crosslinkable chitosan-hyaluronic acid dialdehyde nanoparticles for CD44-targeted siRNA delivery to treat bladder cancer. Bioact. Mater. 2021, 6, 433–446. [Google Scholar] [CrossRef] [PubMed]
- Cannito, S.; Bincoletto, V.; Turato, C.; Pontisso, P.; Scupoli, M.T.; Ailuno, G.; Andreana, I.; Stella, B.; Arpicco, S.; Bocca, C. Hyaluronated and PEGylated Liposomes as a Potential Drug-Delivery Strategy to Specifically Target Liver Cancer and Inflammatory Cells. Molecules 2022, 27, 1062. [Google Scholar] [CrossRef]
- Bi, Y.E.; Zhou, Y.; Wang, M.; Li, L.; Lee, R.J.; Xie, J.; Teng, L. Targeted Delivery of Cordycepin to Liver Cancer Cells Using Transferrin-conjugated Liposomes. Anticancer. Res. 2017, 37, 5207–5214. [Google Scholar] [CrossRef]
- Kengkittipat, W.; Kaewmalun, S.; Khongkow, M.; Iempridee, T.; Jantimaporn, A.; Bunwatcharaphansakun, P.; Yostawonkul, J.; Yata, T.; Phoolcharoen, W.; Namdee, K. Improvement of the multi-performance biocharacteristics of cordycepin using BiloNiosome-core/chitosan-shell hybrid nanocarriers. Colloids Surf. B. Biointerfaces 2021, 197, 111369. [Google Scholar] [CrossRef]
- Marslin, G.; Khandelwal, V.; Franklin, G. Cordycepin Nanoencapsulated in Poly(Lactic-Co-Glycolic Acid) Exhibits Better Cytotoxicity and Lower Hemotoxicity Than Free Drug. Nanotechnol. Sci. Appl. 2020, 13, 37–45. [Google Scholar] [CrossRef]
- Marsup, P.; Yeerong, K.; Neimkhum, W.; Sirithunyalug, J.; Anuchapreeda, S.; To-Anun, C.; Chaiyana, W. Enhancement of Chemical Stability and Dermal Delivery of Cordyceps militaris Extracts by Nanoemulsion. Nanomaterials 2020, 10, 1565. [Google Scholar] [CrossRef]
- Shashidhar, G.M.; Manohar, B. Nanocharacterization of liposomes for the encapsulation of water soluble compounds from Cordyceps sinensis CS1197 by a supercritical gas anti-solvent technique. RSC Adv. 2018, 8, 34634–34649. [Google Scholar] [CrossRef] [Green Version]
- Aramwit, P.; Porasuphatana, S.; Srichana, T.; Nakpheng, T. Toxicity evaluation of cordycepin and its delivery system for sustained in vitro anti-lung cancer activity. Nanoscale Res. Lett. 2015, 10, 152. [Google Scholar] [CrossRef] [Green Version]
- Allahverdiyev, A.M.; Parlar, E.; Dinparvar, S.; Bagirova, M.; Abamor, E. Current aspects in treatment of breast cancer based of nanodrug delivery systems and future prospects. Artif Cells. Nanomed. Biotechnol. 2018, 46, S755–S762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Shi, Q.; Wang, Z.; Gu, Y.; Shen, Y.; Sun, M.; Deng, M.; Zhang, H.; Fang, J.; Zhang, S.; et al. Clinicopathologic correlation of cancer stem cell markers CD44, CD24, VEGF and HIF-1α in ductal carcinoma in situ and invasive ductal carcinoma of breast: An immunohistochemistry-based pilot study. Pathol. Res. Pract. 2011, 207, 505–513. [Google Scholar] [CrossRef] [PubMed]
- Jahan, S.; Karim, M.E.; Chowdhury, E.H. Nanoparticles Targeting Receptors on Breast Cancer for Efficient Delivery of Chemotherapeutics. Biomedicines 2021, 9, 114. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, A.; Waters, A.K.; Kalyan, P.; Achrol, A.S.; Kesari, S.; Yenugonda, V.M. Lipid-polymer hybrid nanoparticles as a next-generation drug delivery platform: State of the art, emerging technologies, and perspectives. Int. J. Nanomed. 2019, 14, 1937–1952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hadinoto, K.; Sundaresan, A.; Cheow, W.S. Lipid–polymer hybrid nanoparticles as a new generation therapeutic delivery platform: A review. Eur. J. Pharm. Biopharm. 2013, 85, 427–443. [Google Scholar] [CrossRef]
- Cheow, W.S.; Hadinoto, K. Factors affecting drug encapsulation and stability of lipid–polymer hybrid nanoparticles. Colloids Surf. B. Biointerfaces 2011, 85, 214–220. [Google Scholar] [CrossRef]
- Hald Albertsen, C.; Kulkarni, J.A.; Witzigmann, D.; Lind, M.; Petersson, K.; Simonsen, J.B. The role of lipid components in lipid nanoparticles for vaccines and gene therapy. Adv. Drug Deliv. Rev. 2022, 188, 114416. [Google Scholar] [CrossRef]
- Animasawun, R.K.; Taresco, V.; Swainson, S.M.E.; Suksiriworapong, J.; Walker, D.A.; Garnett, M.C. Screening and matching polymers with drugs to improve drug incorporation and retention in nanoparticles. Mol. Pharm. 2020, 17, 2083–2098. [Google Scholar] [CrossRef]
- Taresco, V.; Creasey, R.G.; Kennon, J.; Mantovani, G.; Alexander, C.; Burley, J.C.; Garnett, M.C. Variation in structure and properties of poly(glycerol adipate) via control of chain branching during enzymatic synthesis. Polymer 2016, 89, 41–49. [Google Scholar] [CrossRef]
- Fan, Y.; Sahdev, P.; Ochyl, L.J.; Akerberg, J.; Moon, J.J. Cationic liposome–hyaluronic acid hybrid nanoparticles for intranasal vaccination with subunit antigens. J. Control. Release 2015, 208, 121–129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.-J.; Pan, M.-C.; Chang, C.-K.; Chang, S.-W.; Hsieh, C.-W. Optimization of ultrasonic-assisted extraction of cordycepin from Cordyceps militaris using orthogonal experimental design. Molecules 2014, 19, 20808–20820. [Google Scholar] [CrossRef] [Green Version]
- Zeng, W.B.; Yu, H.; Ge, F.; Yang, J.Y.; Chen, Z.H.; Wang, Y.B.; Dai, Y.D.; Adams, A. Distribution of nucleosides in populations of Cordyceps cicadae. Molecules 2014, 19, 6123–6141. [Google Scholar] [CrossRef] [Green Version]
- Long, T.R.; Wongrakpanich, A.; Do, A.-V.; Salem, A.K.; Bowden, N.B. Long-term release of a thiobenzamide from a backbone functionalized poly(lactic acid). Polym. Chem. 2015, 6, 7188–7195. [Google Scholar] [CrossRef] [Green Version]
- Shao, K.; Huang, R.; Li, J.; Han, L.; Ye, L.; Lou, J.; Jiang, C. Angiopep-2 modified PE-PEG based polymeric micelles for amphotericin B delivery targeted to the brain. J. Control. Release 2010, 147, 118–126. [Google Scholar] [CrossRef] [PubMed]
- Shao, K.; Wu, J.; Chen, Z.; Huang, S.; Li, J.; Ye, L.; Lou, J.; Zhu, L.; Jiang, C. A brain-vectored angiopep-2 based polymeric micelles for the treatment of intracranial fungal infection. Biomaterials 2012, 33, 6898–6907. [Google Scholar] [CrossRef]
- Ananda, K.; Nacharaju, P.; Smith, P.K.; Acharya, S.A.; Manjula, B.N. Analysis of functionalization of methoxy-PEG as maleimide-PEG. Anal. Biochem. 2008, 374, 231–242. [Google Scholar] [CrossRef]
- Verheul, R.J.; van der Wal, S.; Hennink, W.E. Tailorable Thiolated Trimethyl Chitosans for Covalently Stabilized Nanoparticles. Biomacromolecules 2010, 11, 1965–1971. [Google Scholar] [CrossRef] [PubMed]
- Suksiriworapong, J.; Taresco, V.; Ivanov, D.P.; Styliari, I.D.; Sakchaisri, K.; Junyaprasert, V.B.; Garnett, M.C. Synthesis and properties of a biodegradable polymer-drug conjugate: Methotrexate-poly(glycerol adipate). Colloids Surf. B. Biointerfaces 2018, 167, 115–125. [Google Scholar] [CrossRef]
- Rao, Y.K.; Fang, S.-H.; Wu, W.-S.; Tzeng, Y.-M. Constituents isolated from Cordyceps militaris suppress enhanced inflammatory mediator’s production and human cancer cell proliferation. J. Ethnopharmacol. 2010, 131, 363–367. [Google Scholar] [CrossRef]
- Zhao, T.-Y.; Lü, G.-P.; Wang, L.-Y.; Zhao, J.; Li, S.-P. HPLC determination of six nucleosides in Cordyceps militaris. Chin. J. Pharm. Anal. 2015, 35, 1078–1082. [Google Scholar]
- Chen, S.-Y.; Ho, K.-J.; Hsieh, Y.-J.; Wang, L.-T.; Mau, J.-L. Contents of lovastatin, γ-aminobutyric acid and ergothioneine in mushroom fruiting bodies and mycelia. LWT 2012, 47, 274–278. [Google Scholar] [CrossRef]
- Pfefferkorn, D.; Pulst, M.; Naolou, T.; Busse, K.; Balko, J.; Kressler, J. Crystallization and melting of poly(glycerol adipate)-based graft copolymers with single and double crystallizable side chains. J. Polym. Sci. Part B Polym. Phys. 2013, 51, 1581–1591. [Google Scholar] [CrossRef]
- Wahab, A.; Favretto, M.E.; Onyeagor, N.D.; Khan, G.M.; Douroumis, D.; Casely-Hayford, M.A.; Kallinteri, P. Development of poly(glycerol adipate) nanoparticles loaded with non-steroidal anti-inflammatory drugs. J. Microencapsul. 2012, 29, 497–504. [Google Scholar] [CrossRef]
- Jacob, P.L.; Brugnoli, B.; Del Giudice, A.; Phan, H.; Chauhan, V.M.; Beckett, L.; Gillis, R.B.; Moloney, C.; Cavanagh, R.J.; Krumins, E.; et al. Poly (diglycerol adipate) variants as enhanced nanocarrier replacements in drug delivery applications. J. Colloid Interface Sci. 2023, 641, 1043–1057. [Google Scholar] [CrossRef]
- Jacob, P.L.; Ruiz Cantu, L.A.; Pearce, A.K.; He, Y.; Lentz, J.C.; Moore, J.C.; Machado, F.; Rivers, G.; Apebende, E.; Fernandez, M.R.; et al. Poly (glycerol adipate) (PGA) backbone modifications with a library of functional diols: Chemical and physical effects. Polymer 2021, 228, 123912. [Google Scholar] [CrossRef]
- Taresco, V.; Tulini, I.; Francolini, I.; Piozzi, A. Polyglycerol adipate-grafted polycaprolactone nanoparticles as carriers for the antimicrobial compound usnic acid. Int. J. Mol. Sci. 2022, 23, 14339. [Google Scholar] [CrossRef] [PubMed]
- Taresco, V.; Suksiriworapong, J.; Creasey, R.; Burley, J.C.; Mantovani, G.; Alexander, C.; Treacher, K.; Booth, J.; Garnett, M.C. Properties of acyl modified poly(glycerol-adipate) comb-like polymers and their self-assembly into nanoparticles. J. Polym. Sci. Part A Polym. Chem. 2016, 54, 3267–3278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taresco, V.; Suksiriworapong, J.; Styliari, I.D.; Argent, R.H.; Swainson, S.M.E.; Booth, J.; Turpin, E.; Laughton, C.A.; Burley, J.C.; Alexander, C.; et al. New N-acyl amino acid-functionalized biodegradable polyesters for pharmaceutical and biomedical applications. RSC Adv. 2016, 6, 109401–109405. [Google Scholar] [CrossRef] [Green Version]
- Paragkumar, N.T.; Edith, D.; Six, J.-L. Surface characteristics of PLA and PLGA films. Appl. Surf. Sci. 2006, 253, 2758–2764. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, Y.; Luo, R.; Chen, S.; Li, X.; Yuan, S.; Wang, J.; Huang, N. In vitro hemocompatibility and cytocompatibility of dexamethasone-eluting PLGA stent coatings. Appl. Surf. Sci. 2015, 328, 154–162. [Google Scholar] [CrossRef]
- Arpicco, S.; Bartkowski, M.; Barge, A.; Zonari, D.; Serpe, L.; Milla, P.; Dosio, F.; Stella, B.; Giordani, S. Effects of the Molecular Weight of Hyaluronic Acid in a Carbon Nanotube Drug Delivery Conjugate. Front. Chem. 2020, 8, 578008. [Google Scholar] [CrossRef]
- Della Sala, F.; Silvestri, T.; Borzacchiello, A.; Mayol, L.; Ambrosio, L.; Biondi, M. Hyaluronan-coated nanoparticles for active tumor targeting: Influence of polysaccharide molecular weight on cell uptake. Colloids Surf. B. Biointerfaces 2022, 210, 112240. [Google Scholar] [CrossRef]
- Qhattal, H.S.S.; Hye, T.; Alali, A.; Liu, X. Hyaluronan Polymer Length, Grafting Density, and Surface Poly(ethylene glycol) Coating Influence in Vivo Circulation and Tumor Targeting of Hyaluronan-Grafted Liposomes. ACS Nano 2014, 8, 5423–5440. [Google Scholar] [CrossRef]
- Zhong, L.; Liu, Y.; Xu, L.; Li, Q.; Zhao, D.; Li, Z.; Zhang, H.; Zhang, H.; Kan, Q.; Sun, J.; et al. Exploring the relationship of hyaluronic acid molecular weight and active targeting efficiency for designing hyaluronic acid-modified nanoparticles. Asian J. Pharm. Sci. 2019, 14, 521–530. [Google Scholar] [CrossRef] [PubMed]
- van Oss, C.J. Chapter Eight-Stability Versus Flocculation of Aqueous Particle Suspensions. In Interface Science and Technology; van Oss, C.J., Ed.; Elsevier: Amsterdam, The Netherlands, 2008; Volume 16, pp. 113–130. [Google Scholar]
- Yoon, S.Y.; Park, S.J.; Park, Y.J. The Anticancer Properties of Cordycepin and Their Underlying Mechanisms. Int. J. Mol. Sci. 2018, 19, 3027. [Google Scholar] [CrossRef] [Green Version]
- Prasad, K.N.; Kumar, B.; Yan, X.-D.; Hanson, A.J.; Cole, W.C. α-Tocopheryl succinate, the most effective form of vitamin E for adjuvant cancer treatment: A review. J. Am. Coll. Nutr. 2003, 22, 108–117. [Google Scholar] [CrossRef]
- Thummarati, P.; Suksiriworapong, J.; Sakchaisri, K.; Nawroth, T.; Langguth, P.; Roongsawang, B.; Junyaprasert, V.B. Comparative study of dual delivery of gemcitabine and curcumin using CD44 targeting hyaluronic acid nanoparticles for cancer therapy. J. Drug Deliv. Sci. Technol. 2022, 77, 103883. [Google Scholar] [CrossRef]
- Kendirgi, F.; Rexer, D.J.; Alcázar-Román, A.R.; Onishko, H.M.; Wente, S.R. Interaction between the shuttling mRNA export factor Gle1 and the nucleoporin hCG1: A conserved mechanism in the export of Hsp70 mRNA. Mol. Biol. Cell 2005, 16, 4304–4315. [Google Scholar] [CrossRef] [Green Version]
- Narvaez, C.J.; LaPorta, E.; Robilotto, S.; Liang, J.; Welsh, J. Inhibition of HAS2 and hyaluronic acid production by 1,25-dihydroxyvitamin D(3) in breast cancer. Oncotarget 2020, 11, 2889–2905. [Google Scholar] [CrossRef]







| Polymers | Mn (g/mol) | Mw/Mn |
|---|---|---|
| PGA | 6300 | 3.1 |
| PGA-CH | 10,600 | 5.4 |
| Mal-PEG-PGA-CH | 9900 | 4.8 |
| PGA-VE | 10,100 | 6.4 |
| Mal-PEG-PGA-VE | 9900 | 4.3 |
| Series | Formulation Code | Weight (mg) of Components Used in Organic Phase | Mean (SD) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| PLGA | PGA-CH/ PGA-VE | Mal-PEG-PGA-CH/ Mal-PEG-PGA-VE | PC | Size (nm) | PDI | ZP (mV) | %Yield | %DL | %EE | ||
| PLGA | CME-PLGA | 18.75 | - | - | 6.25 | 251 (49) | 0.246 (0.039) | –2.2 (1.0) | 78.0 (4.8) | 1.66 (0.27) | 31.1 (6.8) |
| PGA-CH | CME-CH | - | 18.75 | - | 6.25 | 491 *** (290) | 0.428 (0.201) | –5.8 * (0.4) | 81.0 (8.3) | 1.64 (0.13) | 31.6 (2.9) |
| CME-25PEG/CH | - | 14.06 | 4.69 | 6.25 | 220 (21) | 0.227 (0.032) | –6.6 * (1.2) | 80.7 (17.7) | 1.49 (0.35) | 27.8 (3.9) | |
| CME-50PEG/CH | - | 9.38 | 9.38 | 6.25 | 196 (7) | 0.201 (0.021) | –5.4 * (1.0) | 85.8 (5.1) | 1.71 (0.15) | 35.2 (4.9) | |
| PGA-VE | CME-VE | - | 18.75 | - | 6.25 | 969 * (397) | 0.555 * (0.351) | –4.5 (1.8) | 73.8 (0.7) | 1.72 (0.32) | 31.0 (9.2) |
| CME-25PEG/VE | - | 14.06 | 4.69 | 6.25 | 292 ** (17) | 0.450 (0.049) | –5.9 * (1.8) | 75.4 (5.0) | 1.56 (0.22) | 28.8 (9.0) | |
| CME-50PEG/VE | - | 9.38 | 9.38 | 6.25 | 213 ** (28) | 0.312 (0.137) | –4.0 (2.0) | 78.6 (7.6) | 1.62 (0.09) | 31.1 (8.1) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suksiriworapong, J.; Pongprasert, N.; Bunsupa, S.; Taresco, V.; Crucitti, V.C.; Janurai, T.; Phruttiwanichakun, P.; Sakchaisri, K.; Wongrakpanich, A. CD44-Targeted Lipid Polymer Hybrid Nanoparticles Enhance Anti-Breast Cancer Effect of Cordyceps militaris Extracts. Pharmaceutics 2023, 15, 1771. https://doi.org/10.3390/pharmaceutics15061771
Suksiriworapong J, Pongprasert N, Bunsupa S, Taresco V, Crucitti VC, Janurai T, Phruttiwanichakun P, Sakchaisri K, Wongrakpanich A. CD44-Targeted Lipid Polymer Hybrid Nanoparticles Enhance Anti-Breast Cancer Effect of Cordyceps militaris Extracts. Pharmaceutics. 2023; 15(6):1771. https://doi.org/10.3390/pharmaceutics15061771
Chicago/Turabian StyleSuksiriworapong, Jiraphong, Nutthachai Pongprasert, Somnuk Bunsupa, Vincenzo Taresco, Valentina Cuzzucoli Crucitti, Thitapa Janurai, Pornpoj Phruttiwanichakun, Krisada Sakchaisri, and Amaraporn Wongrakpanich. 2023. "CD44-Targeted Lipid Polymer Hybrid Nanoparticles Enhance Anti-Breast Cancer Effect of Cordyceps militaris Extracts" Pharmaceutics 15, no. 6: 1771. https://doi.org/10.3390/pharmaceutics15061771