Enteric Chromosomal Islands: DNA Packaging Specificity and Role of λ-like Helper Phage Terminase
Abstract
:1. Introduction
1.1. DNA Packaging in the λ-like Phages
1.2. Terminase Structure
1.3. Terminase:cos-λ Interactions
1.4. PICI Packaging by λ-like Helper Phages
2. Proposals: Recognition of cos-PICI and the Essential Role of Ters
2.1. The PICI cos
2.2. Rpp Proteins
2.3. Proposals for Rpp-R3 Binding: (1) Location of R3-PICI; (2) Rpp Wing Variation May Reflect Differences in DNA-Binding Specificity
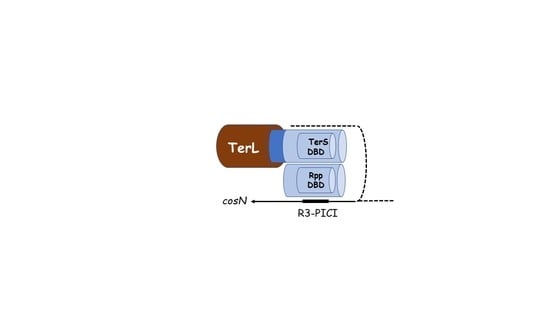
2.4. Hypothesis: The Role of TerS in Hyjacking Is to Recruit TerL to cos-PICI
3. Architecture of the cosN-PICI Nicking Complex
4. Implications for Phage DNA Packaging
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fillol-Salom, A.; Bacarizo, J.; Alqasmi, M.; Ciges-Tomas, J.; Martínez-Rubio, R.; Roszak, A.W.; Cogdell, R.J.; Chen, J.; Marina, A.; Penadés, J.R. Hijacking the Hijackers: Escherichia coli Pathogenicity Islands Redirect Helper Phage Packaging for Their Own Benefit. Mol. Cell 2019, 75, 1020–1030.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fillol-Salom, A.; Martínez-Rubio, R.; Abdulrahman, R.F.; Chen, J.; Davies, R.; Penadés, J.R. Phage-Inducible chromosomal islands are ubiquitous within the bacterial universe. ISME J. 2018, 12, 2114–2128. [Google Scholar] [CrossRef] [PubMed]
- Catalano, C.E.; Cue, D.; Feiss, M. Virus DNA packaging: The strategy used by phage lambda. Mol. Microbiol. 1995, 16, 1075–1086. [Google Scholar] [CrossRef] [PubMed]
- Newcomb, W.W.; Homa, F.L.; Brown, J.C. Involvement of the Portal at an Early Step in Herpes Simplex Virus Capsid Assembly. J. Virol. 2005, 79, 10540–10546. [Google Scholar] [CrossRef] [Green Version]
- Kochan, J.; Carrascosa, J.L.; Murialdo, H. Bacteriophage lambda preconnectors: Purification and structure. J. Mol. Biol. 1984, 174, 433–447. [Google Scholar] [CrossRef]
- Yeo, A.; Feiss, M. Specific interaction of terminase, the DNA packaging enzyme of bacteriophage lambda, with the portal protein of the prohead. J. Mol. Biol. 1995, 245, 141–150. [Google Scholar] [CrossRef]
- Sippy, J.; Feiss, M. Analysis of a mutation affecting the specificity domain for prohead binding of the bacteriophage lambda terminase. J. Bacteriol. 1992, 174, 850–856. [Google Scholar] [CrossRef] [Green Version]
- Davidson, A.R.; Gold, M. Mutations abolishing the endonuclease activity of bacteriophage lambda terminase lie in two distinct regions of the A gene, one of which may encode a “leucine zipper” DNA-binding domain. Virology 1992, 189, 21–30. [Google Scholar] [CrossRef]
- Rao, V.B.; Feiss, M. Mechanisms of DNA Packaging by Large Double-Stranded DNA Viruses. Annu. Rev. Virol. 2015, 2, 351–378. [Google Scholar] [CrossRef] [Green Version]
- Frackman, S.; Siegele, D.; Feiss, M. The terminase of bacteriophage λ: Functional domains for cosB binding and multimer assembly. J. Mol. Biol. 1985, 183, 225–238. [Google Scholar] [CrossRef]
- Bain, D.L.; Berton, N.; Ortega, M.; Baran, J.; Yang, Q.; Catalano, C.E. Biophysical Characterization of the DNA Binding Domain of gpNu1, a Viral DNA Packaging Protein. J. Biol. Chem. 2001, 276, 20175–20181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Araya, P.; Rosemblatt, M.; Valenzuela, P.; Murialdo, H. The bacteriophage lambda DNA packaging enzyme: Identification of four structural domains of the gpNu1 subunit using limited proteolysis. Biol. Res. 2001, 34, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Leavitt, J.C.; Gilcrease, E.B.; Wilson, K.; Casjens, S.R. Function and horizontal transfer of the small terminase subunit of the tailed bacteriophage Sf6 DNA packaging nanomotor. Virology 2013, 440, 117–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, A.; Bhardwaj, A.; Datta, P.; Lander, G.C.; Cingolani, G. Small Terminase Couples Viral DNA Binding to Genome-Packaging ATPase Activity. Structure 2012, 20, 1403–1413. [Google Scholar] [CrossRef] [Green Version]
- Frackman, S.; Siegele, D.A.; Feiss, M. A functional domain of bacteriophage lambda terminase for prohead binding. J. Mol. Biol. 1984, 180, 283–300. [Google Scholar] [CrossRef]
- Arens, J.S.; Duffy, C.; Feiss, M. Acidic residues and a predicted, highly conserved α-helix are critical for the endonuclease/strand separation functions of bacteriophage λ’s TerL. Mol. Microbiol. 2019, 112, 1483–1498. [Google Scholar] [CrossRef]
- Maluf, N.K.; Gaussier, H.; Bogner, E.; Feiss, M.; Catalano, C.E. Assembly of Bacteriophage Lambda Terminase into a Viral DNA Maturation and Packaging Machine. Biochemistry 2006, 45, 15259–15268. [Google Scholar] [CrossRef]
- Vahanian, N.; Oh, C.S.; Sippy, J.; Feiss, M. Natural history of a viral cohesive end site: cosN of the lambda-like phages. Virology 2017, 509, 140–145. [Google Scholar] [CrossRef]
- Feiss, M.; Min, J.Y.; Sultana, S.; Patel, P.; Sippy, J. DNA Packaging Specificity of Bacteriophage N15 with an Excursion into the Genetics of a Cohesive End Mismatch. PLoS ONE 2015, 10, e0141934. [Google Scholar] [CrossRef] [Green Version]
- Feiss, M.; Geyer, H.; Klingberg, F.; Moreno, N.; Forystek, A.; Maluf, N.K.; Sippy, J. Novel DNA packaging recognition in the unusual bacteriophage N15. Virology 2015, 482, 260–268. [Google Scholar] [CrossRef] [Green Version]
- Alanazi, A.M.; Neidle, E.L.; Momany, C. The DNA-binding domain of BenM reveals the structural basis for the recognition of a T-N-11-A sequence motif by LysR-type transcriptional regulators. Acta Crystallogr. Sect. D-Struct. Biol. 2013, 69, 1995–2007. [Google Scholar] [CrossRef] [PubMed]
- de Beer, T.; Fang, J.; Ortega, M.; Yang, Q.; Maes, L.; Duffy, C.; Berton, N.; Sippy, J.; Overduin, M.; Feiss, M.; et al. Insights into specific DNA recognition during the assembly of a viral genome packaging machine. Mol. Cell 2002, 9, 981–991. [Google Scholar] [CrossRef]
- Pabo, C.O.; Sauer, R.T. Protein-DNA recognition. Annu. Rev. Biochem. 1984, 53, 293–321. [Google Scholar] [CrossRef] [PubMed]
- Cue, D.; Feiss, M. Genetic-Analysis of cosB, the Binding-Site for Terminase, the DNA Packaging Enzyme of Bacteriophage-Lambda. J. Mol. Biol. 1992, 228, 58–71. [Google Scholar] [CrossRef]
- Miller, G.; Feiss, M. The bacteriophage lambda cohesive end site: Isolation of spacing/substitution mutations that result in dependence on Escherichia coli integration host factor. Mol. Gen. Genet. 1988, 212, 157–165. [Google Scholar] [CrossRef]
- Higgins, R.R.; Becker, A. The lambda terminase enzyme measures the point of its endonucleolytic attack 47 +/- 2 bp away from its site of specific DNA binding, the R site. Embo J. 1994, 13, 6162–6171. [Google Scholar] [CrossRef]
- Sippy, J.; Patel, P.; Vahanian, N.; Sippy, R.; Feiss, M. Genetics of critical contacts and clashes in the DNA packaging specificities of bacteriophages lambda and 21. Virology 2015, 476, 115–123. [Google Scholar] [CrossRef] [Green Version]
- Hwang, Y.; Feiss, M. Mutations Affecting Lysine-35 of gpNu1, the Small Subunit of Bacteriophage λ Terminase, Alter the Strength and Specificity of Holoterminase Interactions with DNA. Virology 1997, 231, 218–230. [Google Scholar] [CrossRef] [Green Version]
- Smith, M.P.; Feiss, M. Sites and gene products involved in lambdoid phage DNA packaging. J. Bacteriol. 1993, 175, 2393–2399. [Google Scholar] [CrossRef] [Green Version]
- Punekar, A.S.; Porter, J.; Carr, S.B.; Phillips, S.E.V. Structural basis for DNA recognition by the transcription regulator MetR. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2016, 72, 417–426. [Google Scholar] [CrossRef] [Green Version]
- de Beer, T.; Ortega, M.; Berton, N.; Yang, Q.; Overduin, M.; Catalano, C.E. Assignment of the 1H, 13C, and 15N resonances of the DNA binding domain of gpNu1, a genome packaging protein from bacteriophage lambda. J. Biomol. NMR 2000, 18, 69–70. [Google Scholar] [CrossRef] [PubMed]
- Becker, A.; Murialdo, H. Bacteriophage lambda DNA: The beginning of the end. J. Bacteriol. 1990, 172, 2819–2824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maluf, N.K.; Yang, Q.; Catalano, C.E. Self-Association properties of the bacteriophage lambda terminase holoenzyme: Implications for the DNA packaging motor. J. Mol. Biol. 2005, 347, 523–542. [Google Scholar] [CrossRef] [PubMed]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Murialdo, H.; Feiss, M. Enteric Chromosomal Islands: DNA Packaging Specificity and Role of λ-like Helper Phage Terminase. Viruses 2022, 14, 818. https://doi.org/10.3390/v14040818
Murialdo H, Feiss M. Enteric Chromosomal Islands: DNA Packaging Specificity and Role of λ-like Helper Phage Terminase. Viruses. 2022; 14(4):818. https://doi.org/10.3390/v14040818
Chicago/Turabian StyleMurialdo, Helios, and Michael Feiss. 2022. "Enteric Chromosomal Islands: DNA Packaging Specificity and Role of λ-like Helper Phage Terminase" Viruses 14, no. 4: 818. https://doi.org/10.3390/v14040818