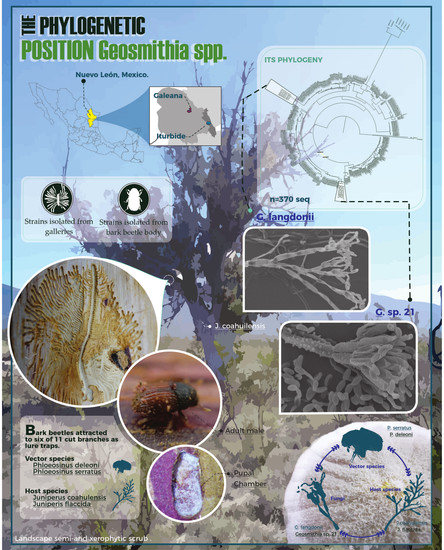
Phylogenetic Position of Geosmithia spp. (Hypocreales) Living in Juniperus spp. Forests (Cupressaceae) with Bark Beetles of Phloeosinus spp. (Scolytinae) from the Northeast of Mexico
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Cultural and Morphological Characteristics
2.3. DNA Extraction, Amplification, and Sequencing
2.4. Phylogenetic Analyses
3. Results
3.1. Phylogenetic Analysis
3.2. Morphological Characterization
4. Discussion
4.1. Identity of Geosmithia Strains
4.2. Geographic, Bark Beetle Vector, and Host Tree Records
4.3. Geosmithia Diversity
4.4. Entomochory in Geosmithia Strains
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Six, D.L.; Wingfield, M.J. The Role of Phytopathogenicity in Bark Beetle–Fungus Symbioses: A Challenge to the Classic Paradigm. Annu. Rev. Entomol. 2011, 56, 255–272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skelton, J.; Jusino, M.A.; Carlson, P.S.; Smith, K.; Banik, M.T.; Lindner, D.L.; Palmer, J.M.; Hulcr, J. Relationships among Wood-Boring Beetles, Fungi, and the Decomposition of Forest Biomass. Mol. Ecol. 2019, 28, 4971–4986. [Google Scholar] [CrossRef] [PubMed]
- Harrington, T.C. Ecology and Evolution of Mycophagous Bark Beetles and Their Fungal Partners. Insect-Fungal Assoc. Ecol. Evol. 2005, 257–291. [Google Scholar]
- Kok, L.T.; Norris, D.M.; Chu, H.M. Sterol metabolism as a basis for a mutualistic symbiosis. Nature 1970, 225, 661–662. [Google Scholar] [CrossRef] [PubMed]
- Hsiau, P.T.W.; Harrington, T.C. Phylogenetics and Adaptations of Basidiomycetous Fungi Fed upon by Bark Beetles (Coleoptera: Scolytidae). Symbiosis 2003, 34, 111–131. [Google Scholar]
- Wingfield, M.J.; van Wyk, P.S. A new species of Ophiostoma from Protea infructescences in South Africa. Mycol. Res. 1993, 97, 709–716. [Google Scholar] [CrossRef]
- Kirisits, T. Fungal associates of European bark beetles with special emphasis on the ophiostomatoid fungi In Bark and Wood Boring Insects in Living Trees in Europe: A Synthesis; Lieutier, F., Day, K.R., Battisti, A., Grégoire, J.C., Evans, H.F., Eds.; Kluwer Acad: Dordrecht, The Netherlands, 2004; pp. 181–236. [Google Scholar]
- Linnakoski, R.; Wilhelm de Beer, Z.B.; Niemelä, P.; Wingfield, M.J. Associations of Conifer-Infesting Bark Beetles and Fungi in Fennoscandia. Insects 2012, 3, 200–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benjamin, R.K.; Blackwell, M.E.R.E.D.I.T.H.; Chapela, I.H.; Humber, R.A.; Jones, K.G.; Klepzig, K.D.; SPATAFORA, J.W. Insect and Other Arthropod-Associated Fungi. Biodiversity of Fungi: Inventory and Monitoring Methods; Elsevier: Burlington, MA, USA, 2004; pp. 395–434. [Google Scholar]
- Jankowiak, R.; Kolařík, M.; Bilański, P. Association of Geosmithia Fungi (Ascomycota: Hypocreales) with Pine- and Spruce-Infesting Bark Beetles in Poland. Fungal Ecol. 2014, 11, 71–79. [Google Scholar] [CrossRef]
- Kolařík, M.; Jankowiak, R. Vector Affinity and Diversity of Geosmithia Fungi Living on Subcortical Insects Inhabiting Pinaceae Species in Central and Northeastern Europe. Microb. Ecol. 2013, 66, 682–700. [Google Scholar] [CrossRef]
- Dori-Bachash, M.; Avrahami-Moyal, L.; Protasov, A.; Mendel, Z.; Freeman, S. The Occurrence and Pathogenicity of Geosmithia spp. and Common Blue-Stain Fungi Associated with Pine Bark Beetles in Planted Forests in Israel. Eur. J. Plant. Pathol. 2015, 143, 627–639. [Google Scholar] [CrossRef]
- Kolařík, M.; Kubátová, A.; Pažoutová, S.; Šrůtka, P. Morphological and Molecular Characterisation of Geosmithia putterillii, G. pallida Comb. Nov. and G. flava sp. Nov., Associated with Subcorticolous Insects. Mycol. Res. 2004, 108, 1053–1069. [Google Scholar] [CrossRef]
- Kolařík, M.; Kubátová, A.; Čepička, I.; Pažoutová, S.; Šrůtka, P. A Complex of Three New White-Spored, Sympatric, and Host Range Limited Geosmithia Species. Mycol. Res. 2005, 109, 1323–1336. [Google Scholar] [CrossRef] [Green Version]
- Kolařík, M.; Kostovčík, M.; Pažoutová, S. Host Range and Diversity of the Genus Geosmithia (Ascomycota: Hypocreales) Living in Association with Bark Beetles in the Mediterranean Area. Mycol. Res. 2007, 111, 1298–1310. [Google Scholar] [CrossRef]
- Kolařík, M.; Kubátová, A.; Hulcr, J.; Pažoutová, S. Geosmithia Fungi Are Highly Diverse and Consistent Bark Beetle Associates: Evidence from Their Community Structure in Temperate Europe. Microb. Ecol. 2008, 55, 65–80. [Google Scholar] [CrossRef] [PubMed]
- Kolařík, M.; Freeland, E.; Utley, C.; Tisserat, N. Geosmithia morbida sp. nov., a New Phytopathogenic Species Living in Symbiosis with the Walnut Twig Beetle (Pityophthorus juglandis) on Juglans in USA. Mycologia 2011, 103, 325–332. [Google Scholar] [CrossRef] [PubMed]
- Kolařík, M.; Hulcr, J.; Kirkendall, L.R. New Species of Geosmithia and Graphium Associated with Ambrosia Beetles in Costa Rica. Czech. Mycol. 2015, 67, 29–35. [Google Scholar] [CrossRef]
- Kolařík, M.; Hulcr, J.; Tisserat, N.; De Beer, W.; Kostovčík, M.; Kolaříková, Z.; Seybold, S.J.; Rizzo, D.M. Geosmithia Associated with Bark Beetles and Woodborers in the Western USA: Taxonomic Diversity and Vector Specificity. Mycologia 2017, 109, 185–199. [Google Scholar] [CrossRef] [Green Version]
- Kolařík, M.; Kirkendall, L.R. Evidence for a New Lineage of Primary Ambrosia Fungi in Geosmithia Pitt (Ascomycota: Hypocreales). Fungal Biol. 2010, 114, 676–689. [Google Scholar] [CrossRef]
- Machingambi, N.M.; Roux, J.; Dreyer, L.L.; Roets, F. Bark and Ambrosia Beetles (Curculionidae: Scolytinae), Their Phoretic Mites (Acari) and Associated Geosmithia Species (Ascomycota: Hypocreales) from Virgilia Trees in South Africa. Fungal Biol. 2014, 118, 472–483. [Google Scholar] [CrossRef] [Green Version]
- Pepori, A.L.; Kolařík, M.; Bettini, P.P.; Vettraino, A.M.; Santini, A. Morphological and Molecular Characterisation of Geosmithia Species on European Elms. Fungal Biol. 2015, 119, 1063–1074. [Google Scholar] [CrossRef]
- Pitt, J.I. Geosmithia Gen. Nov. for Penicillium lavendulum and Related Species. Can. J. Bot. 1979, 57, 2021–2030. [Google Scholar] [CrossRef]
- Lin, Y.; Shih, H.; Huang, Y.; Lin, C.; Chen, C. Two Species of Beetle-Associated Geosmithia in Taiwan. Fungal Sci. 2016, 31, 29–36. [Google Scholar]
- Crous, P.W.; Luangsa-Ard, J.J.; Wingfield, M.J.,; Carnegie, A.J.; Hernández-Restrepo, M.; Lombard, L.; Roux, J.; Barreto, R.W.; Baseia, I.G.; Cano-Lira, J.F.; et al. Fungal Planet description sheets: 785–867. Pers. Mol. Phylogeny Evol. Fungi 2018, 41, 238. [Google Scholar] [CrossRef] [PubMed]
- Pitt, J.I.; Hocking, A.D. Fungi and Food Spoilage; Springer: New York, NY, USA, 2009; pp. 1–519. [Google Scholar] [CrossRef]
- McPherson, B.A.; Erbilgin, N.; Bonello, P.; Wood, D.L. Fungal Species Assemblages Associated with Phytophthora Ramorum-Infected Coast Live Oaks Following Bark and Ambrosia Beetle Colonization in Northern California. For. Ecol. Manag. 2013, 291, 30–42. [Google Scholar] [CrossRef]
- Hulcr, J.; Dunn, R.R. The Sudden Emergence of Pathogenicity in Insect-Fungus Symbioses Threatens Naive Forest Ecosystems. Proc. R. Soc. B Biol. Sci. 2011, 278, 2866–2873. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.T.; Skelton, J.; Johnson, A.J.; Kolařík, M.; Hulcr, J. Geosmithia Species in Southeastern USA and Their Affinity to Beetle Vectors and Tree Hosts. Fungal Ecol. 2019, 39, 168–183. [Google Scholar] [CrossRef]
- Schedl, K.E. Bestimmungstabellen der palaearktischen Borkenkäfer. Teil IV. Die Gattung Ips De Geer. Mitt. Forstl. Bundes-Vers. Mariabrunn 1950, 46, 67–88. [Google Scholar]
- Wood, S.L. The bark and ambrosia beetles of North and Central America (Coleoptera: Scolytidae), a taxonomic monograph. Great Basin Nat. 1982, 1086. Available online: https://www.cabdirect.org/cabdirect/abstract/19820595039 (accessed on 10 August 2020).
- Wood, S.L.; Bright, D.E., Jr. A catalog of Scolytidae and Platypodidae (Coleoptera), Part 2. Taxonomic Index. Great Basin Nat. Mem. 1992, 13, 1–1553. [Google Scholar]
- Knížek, M. Scolytinae. In Catalogue of Palaearctic Coleoptera; Löbl, I., Smetana, A., Eds.; Apollo Books: Stenstrup, Denmark, 2011; Volume 7, pp. 86–87, 204–251. [Google Scholar]
- Pfeffer, A. Zentral- und westpaläarktische Borken- und Kernkäfer (Coleoptera: Scolytidae, Platypodidae); Pro Entomologia: Basel, Switzerland, 1995. [Google Scholar]
- Moraal, L.G. Infestations of the cypress bark beetles Phloeosinus rudis, P. bicolor and P. thujae in The Netherlands (Coleoptera: Curculionidae: Scolytinae). Entomol. Ber. 2010, 70, 140–145. [Google Scholar]
- FAO (Food and Agriculture Organization of the United Nations). Global Review of Forest Pests and Diseases; FAO Forestry Paper: Rome, Italy, 2009; Volume 156, p. 222. ISBN 978-92-5-106208-1. [Google Scholar]
- Deng, J.; Guo, Y.; Cheng, Z.; Lu, C.; Huang, X. The Prevalence of Single-Specimen/Locality Species in Insect Taxonomy: An Empirical Analysis. Diversity 2019, 11, 106. [Google Scholar] [CrossRef] [Green Version]
- Morrone, J.J. Biogeographic Regionalization of the Mexican Transition Zone. In The Mexican Transition Zone; Springer: Cham, Switzerland, 2020. [Google Scholar]
- Villaseñor, J.L. Checklist of the native vascular plants of Mexico. Rev. Mex. Biodivers. 2016, 87, 559–902. [Google Scholar] [CrossRef] [Green Version]
- Estrada-Castillón, E.; Villarreal-Quintanilla, J.A.; Salinas-Rodríguez, M.M.; Encina-Domínguez, J.A.; Cantú-Ayala, C.M.; González-Rodríguez, H.; Jiménez-Pérez, J. Coníferas de Nuevo León, México; Universidad Autónoma de Nuevo León: Nuevo León, Mexico, 2014. [Google Scholar]
- Martínez-Torres, A.; Gutiérrez-Ambrocio, S.; Heredia-del-Orbe, P.; Villa-Tanaca, L.; Hernández-Rodríguez, C. Inferring the Role of Microorganisms in Water Kefir Fermentations. Int. J. Food Sci. Technol. 2017, 52, 559–571. [Google Scholar] [CrossRef]
- Harris, J.L. Modified method for fungal slide culture. J. Clin. Microbiol. 1986, 24, 460–461. [Google Scholar] [CrossRef] [Green Version]
- Aylmore, R.C.; Todd, N.K. A microculture chamber and improved method for combined light and electron microscopy of filamentous fungi. J. Microbiol. Methods 1984, 2, 317–322. [Google Scholar] [CrossRef]
- Cole, G.T.; Nag Raj, T.R.; Kendrick, W.B. A simple technique for time-lapse photomicrography of microfungi in plate culture. Mycologia 1969, 61, 726–730. [Google Scholar] [CrossRef]
- Hernández-García, J.A.; Briones-Roblero, C.I.; Rivera-Orduña, F.N.; Zúñiga, G. Revealing the gut bacteriome of Dendroctonus bark beetles (Curculionidae: Scolytinae): Diversity, core members and co-evolutionary patterns. Sci. Rep. 2017, 7, 13864. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. 1990, 315–322. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [Green Version]
- Guindon, S.; Gascuel, O. PhyML: A simple, fast and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jankowiak, R.; Bilański, P. Geosmithia Species Associated with Fir-Infesting Beetles in Poland. Acta Mycol. 2018, 53, 1–10. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, L.T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stöver, B.C.; Müller, K.F. TreeGraph 2: Combining and visualizing evidence from different phylogenetic analyses. BMC Bioinform. 2010, 11, 7. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evo.l. 2018, 3, 1547–1549. [Google Scholar] [CrossRef]
- Skouboe, P.; Frisvad, J.C.; Taylor, J.W.; Lauritsen, D.; Boysen, M.; Rossen, L. Phylogenetic analysis of nucleotide sequences from the ITS region of terverticillate Penicillium species. Mycol. Res. 1999, 103, 873–881. [Google Scholar] [CrossRef]
- Huang, Y.T.; Kolařík, M.; Kasson, M.T.; Hulcr, J. Two new Geosmithia species in G. pallida species complex from bark beetles in eastern USA. Mycologia 2017, 109, 790–803. [Google Scholar] [CrossRef]
- Pepori, A.L.; Bettini, P.P.; Comparini, C.; Sarrocco, S.; Bonini, A.; Frascella, A.; Ghelardini, L.; Scala, A.; Vannacci, G.; Santini, A. Geosmithia-Ophiostoma: A New Fungus-Fungus Association. Microb. Ecol. 2018, 75, 632–646. [Google Scholar] [CrossRef] [Green Version]







| Acronym | Locality | Longitude W | Latitude N | Host | Vector |
|---|---|---|---|---|---|
| JC1 | Area one, Galeana | 24°51′34.8″ | −100°21′37.2″ | Juniperus coahuilensis | Phloeosinus serratus |
| JC2 | 24°51′17.9″ | −100°21′46.0″ | |||
| JC3 | 24°51′18.3″ | −100°21′36.0″ | |||
| JC4 | 24°51′18.1″ | −100°21′42.9″ | |||
| JF1 | Area two, Iturbide | 24°41′50.2″ | −99°52′8.9″ | Juniperus flaccida | Phloeosinus deleoni |
| JF2 | 24°41′50.5″ | −99°52′9.5″ | |||
| JF3 | 24°41′50.4″ | −99°52′9.8″ | |||
| JF4 | 24°41′51.5″ | −99°52′9.6″ | |||
| JF5 | 24°41′51.8″ | −99°52′10.2″ | |||
| JF6 | 24°41′50.0″ | −99°52′9.3″ | |||
| JF7 | 24°10′4.9″ | −100°4′5.6″ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Juan Alfredo, H.-G.; Gerardo, C.-R.; Nallely Guadalupe, A.-O.; Lourdes, V.-T.; César, H.-R.; Francisco, A.-T. Phylogenetic Position of Geosmithia spp. (Hypocreales) Living in Juniperus spp. Forests (Cupressaceae) with Bark Beetles of Phloeosinus spp. (Scolytinae) from the Northeast of Mexico. Forests 2020, 11, 1142. https://doi.org/10.3390/f11111142
Juan Alfredo H-G, Gerardo C-R, Nallely Guadalupe A-O, Lourdes V-T, César H-R, Francisco A-T. Phylogenetic Position of Geosmithia spp. (Hypocreales) Living in Juniperus spp. Forests (Cupressaceae) with Bark Beetles of Phloeosinus spp. (Scolytinae) from the Northeast of Mexico. Forests. 2020; 11(11):1142. https://doi.org/10.3390/f11111142
Chicago/Turabian StyleJuan Alfredo, Hernández-García, Cuellar-Rodríguez Gerardo, Aguirre-Ojeda Nallely Guadalupe, Villa-Tanaca Lourdes, Hernández-Rodríguez César, and Armendáriz-Toledano Francisco. 2020. "Phylogenetic Position of Geosmithia spp. (Hypocreales) Living in Juniperus spp. Forests (Cupressaceae) with Bark Beetles of Phloeosinus spp. (Scolytinae) from the Northeast of Mexico" Forests 11, no. 11: 1142. https://doi.org/10.3390/f11111142