2. Materials and Methods
LANDIS-II NECN and PPA-SiBGC were parameterized for two forested sites in the eastern United States, Harvard Forest, Massachusetts and Jones Ecological Research Center, Georgia. At the HF site, we focus on Little Prospect Hill and the EMS EC flux tower (HF-EMS). At the JERC site, we focus on the mesic zone and RD EC flux tower (JERC-RD). Both sites provided local EC and meteorological measurements to conduct this study. Plots of EC flux and meteorological tower measurements for both sites are located in
Appendix A (
Figure A1,
Figure A2,
Figure A3 and
Figure A4); maps of both sites are provided in the site descriptions.
Both models were parameterized using data available for each site, including local (i.e., field measurements) and general information sources (e.g., species compendiums and other published sources). As these empirical or observational values were used to parameterize both models, further model calibration (i.e., parameter tuning) was not necessary. This is because tuning parameters away from measured values to improve model performance, or defining a separate set of tuning parameters, is known to produce model over-fitting (i.e., reduced generality) and thus false improvements in model accuracy through reduced parsimony [
89]. We explicitly avoided this practice, as it is only appropriate when fitting empirical growth-and-yield models such as Prognosis, also known as the Forest Vegetation Simulator (FVS) [
18,
19]. All model parameters are provided in
Appendix B (
Table A1,
Table A2,
Table A3,
Table A4,
Table A5,
Table A6,
Table A7,
Table A8,
Table A9,
Table A10,
Table A11,
Table A12,
Table A13,
Table A14,
Table A15,
Table A16,
Table A17,
Table A18,
Table A19,
Table A20,
Table A21,
Table A22,
Table A23,
Table A24,
Table A25,
Table A26,
Table A27,
Table A28,
Table A29 and
Table A30). We close the methodology section with descriptions of the metrics, models, and criteria used in the intercomparisons.
2.1. Model Descriptions
In the following sections, we provide a brief overview of the two forest ecosystem models used in this intercomparison study. For detailed information on each model, readers are encouraged to refer to the original publications.
2.1.1. LANDIS-II NECN
The LANDIS-II model is an extension of the original LANdscape DIsturbance and Succession (LANDIS) model [
90,
91,
92] into a modular software framework [
87]. Specifically, LANDIS-II is a model core containing basic state information that interfaces or communicates with external user-developed models known as “extensions” using a combination of object-oriented and modular design. This design makes LANDIS-II a modeling framework rather than a model. The LANDIS family of models, which also includes LANDIS PRO [
93] and Fin-LANDIS [
94,
95], are stochastic hybrid models [
29] based on the vital attributes/fuzzy systems approach of the LANDSIM model genre [
96]. This genre borrows heavily from cellular automata [
97] and thus Markov Chains by applying simple heuristic rule-based systems, in the form of vital attributes, across two-dimensional grids.
Models of the LANDSIM genre focus on landscape-scale processes and assume game-theoretic vital attribute controls over successional trajectories following disturbance [
98]. The LANDSIM model genre is thus a reasonable match for the classical forest fire model [
99], given its local two-dimensional cellular basis. In contrast to the original LANDIS model, LANDIS-II is implemented in Microsoft C# rather than ISO C++98 [
100], simplifying model development in exchange for a proprietary single-vendor software stack [
87].
The latest version of LANDIS-II (v7) supports Linux through use of the Microsoft .NET Core developer platform. The modular design of LANDIS-II is intended to simplify the authorship and interaction of user-provided libraries for succession and disturbance. The centralized model core stores basic landscape and species state information and acts as an interface between succession and disturbance models. While there have been numerous forest landscape models over the years [
21,
22,
23,
24,
25], the LANDIS family of models has enjoyed notable longevity and is currently united under the LANDIS-II Foundation. Part of its longevity is attributable to the prioritization of model functionality over realism in order to appeal to application-minded managers seeking a broad array of functionality.
The Net Ecosystem Carbon and Nitrogen (NECN) model [
101] is a simplified variant of the classical Century model [
36,
37]. The original ten soil layers in Century have been replaced by a single soil layer, with functions for growth and decay borrowed directly from Century v4.5. The NECN succession model
Figure 1 is thus a process-based model that simulates C and N dynamics along the plant-soil continuum at a native monthly timestep.
Atmospheric effects are included through monthly climate (i.e., temperature maxima, minima, means, and standard deviations, and precipitation means and standard deviations). Explicit geometric representation of tree canopies is forgone in favor of bounded statistical growth models based theoretically on Liebig’s Law of the Minimum. Functions for growth, mortality, and decay are adopted from Century [
36] while hydrology is based on the simple bucket model [
102]. The regeneration function is the only new process in NECN and is also based on Liebig’s Law. For a detailed description of the NECN model, readers may refer to the original model publication [
101]. Parameterization of the LANDIS-II model for both sites was based on updating parameters used in recent [
103,
104,
105,
106] and ongoing (Flanagan et al., in review) work.
2.1.2. PPA-SiBGC
The PPA-SiBGC model belongs to the SORTIE-PPA family of models [
46,
49] within the SAS-PPA model genre, based on a simple and analytically tractable approximation of the classical SORTIE gap model [
107,
108]. The Perfect Plasticity Approximation, or PPA [
46,
47], was derived from the dual assumptions of perfect crown plasticity (e.g., space-filling) and phototropism (e.g., stem-leaning), both of which were supported in empirical and modeling studies [
49]. The discovery of the PPA was rooted in extensive observational and in silico research [
46]. The PPA model was designed to overcome the most computationally challenging aspects of gap models in order to facilitate model scaling from the landscape to global scale.
The PPA and its predecessor, the size-and-age structured (SAS) equations [
43,
109], are popular model reduction techniques employed in current state-of-the-art terrestrial biosphere models [
28]. The PPA model can be thought of metaphorically as Navier–Stokes equations of forest dynamics, capable of modeling individual tree population dynamics with a one-dimensional von Foerster partial differential equation [
46]. The simple mathematical foundation of the PPA model is provided in Equation (
1).
where
k is the number of species,
j is the species index,
is the density of species
j at height
z,
is the projected crown area of species
j at height
z, and
is the derivative of height. In other words, we discard the spatial location of individual trees and calculate the height at which the integral of tree crown area is equal to the ground area of the stand. This height is known as the theoretical
height, which segments trees into overstory and understory classes [
46].
The segmentation of the forest canopy into understory and overstory layers allows for separate coefficients or functions for growth, mortality, and fecundity to be applied across strata, whose first moment accurately approximates the dynamics of individual-based forest models. Recent studies have shown that the PPA model faithfully reduces the dynamics of the more recent neighborhood dynamics (ND) SORTIE-ND gap model [
110] and is capable of accurately capturing forest dynamics [
111,
112].
In this work, we applied a simple biogeochemistry variant of the SORTIE-PPA model, PPA-SiBGC (Erickson and Strigul, in review)
Figure 2.
Empirical observations were relied upon for the C and N content of tree species compartments. Stoichiometric relations were used to estimate N from C, based on empirical measurements provided for both sites. All values were calculated directly from observations. Previously published equations [
113] and parameters [
114] were used to model crown allometry. Together with inventory data, general biomass equations were used to estimated dry weight mass (
) for tree stems, branches, leaves, and, fine and coarse roots [
115]. Carbon content was assumed to be 50% of dry mass, generally supported by data. Monthly soil respiration was modeled using the approach of Raich et al. [
116], while soil organic C was modeled using the simple generalized approach of Domke et al. [
117]. Species- and stratum-specific parameters for growth, mortality, and fecundity were calculated directly from field data for both sites. Net ecosystem exchange, or NEE, was modeled as
following previous studies, which note associated challenges in connecting field and flux tower measurements [
118,
119]. Here, ANPP, or annual net primary production, is the total site biomass increment adjusted for the C fraction. This is necessary given the current field-measurement basis of the PPA, which may be replaced by LiDAR measurements and/or process models in future work.
2.2. Site Descriptions
In the following sections, we describe the two forested sites on the East Coast of the United States: HF-EMS and the JERC-RD. A critical factor in the selection of the sites was the availability of eddy covariance flux tower data needed to validate NEE in the models.
2.2.1. HF-EMS
The HF-EMS EC flux tower is located within the Little Prospect Hill tract of Harvard Forest (42.538° N, 72.171° W, 340 m elevation) in Petersham, Massachusetts, approximately 100 km from the city of Boston [
88]. A map of the site is shown in
Figure 3. The tower has been recording NEE, heat, and meteorological measurements since 1989, with continuous measurements since 1991, making it the longest-running eddy covariance measurement system in the world. The site is currently predominantly deciduous broadleaf second-growth forests approximately 75–95 years in age, based on previous estimates [
120]. Soils at Harvard Forest originate from sandy loam glacial till and are reported to be mildly acidic [
88].
The site is dominated by red oak (
Quercus rubra L.) and red maple (
Acer rubrum L.) stands, with sporadic stands of Eastern hemlock (
Tsuga canadensis (L.) Carrière), white pine (
Pinus strobus L.), and red pine (
Pinus resinosa Ait.). When the site was established, it contained 100 Mg C
in live aboveground woody biomass [
120]. As noted by Urbanski et al. [
88], approximately 33% of red oak stands were established prior to 1895, 33% prior to 1930, and 33% before 1940. A relatively hilly and undisturbed forest (since the 1930s) extends continuously for several
around the tower. In 2000, harvest operations removed 22.5 Mg C
of live aboveground woody biomass about 300 m S-SE from the tower, with little known effect on the flux tower measurements. The 40 biometric plots were designated via stratified random sampling within eight 500 m transects Urbanski et al. [
88]. The HF-EMS tower site currently contains 34 biometric plots at 10 m radius each, covering 10,681
, or approximately one hectare, in area. Summary statistics for the EMS tower site for the year 2002 are provided in
Table 1.
A table of observed species abundances for the year 2002 are provided in
Table 2, using tree species codes from the USDA PLANTS database (
https://plants.usda.gov).
Previous research at the EMS EC flux tower site found unusually high rates of ecosystem respiration in winter and low rates in mid-to-late summer compared to other temperate forests [
122]. While the mechanisms behind these observed patterns remains poorly understood, this observation is outside the scope of the presented research. Between 1992 and 2004, the site acted as a net carbon sink, with a mean annual uptake rate of 2.5 Mg C ha
year
. Aging dominated the site characteristics, with a 101–115 Mg C ha
increase in biomass, comprised predominantly of growth of red oak (
Quercus rubra). The year 1998 showed a sharp decline in net ecosystem exchange (NEE) and other metrics, recovering thereafter [
88]. As Urbanski et al. [
88] note of the Integrated Biosphere Simulator 2 (IBIS2) and similar models at the time, “the drivers of interannual and decadal changes in NEE are long-term increases in tree biomass, successional change in forest composition, and disturbance events, processes not well represented in current models.” The two models used in the intercomparison study, a SORTIE-PPA [
46,
47] variant and LANDIS-II with NECN succession [
87,
101], are intended to directly address these model shortcomings.
2.2.2. JERC-RD
Jones Ecological Research Center at Ichauway is located near Newton, Georgia, USA (31° N, 84° W, 25–200 m elevation). A map of the JERC-RD flux tower with landcover classes is shown in
Figure 4. The site falls within the East Gulf Coastal Plain and consists of flat to rolling land sloping to the southwest. The region is characterized by a humid subtropical climate with temperatures ranging from 5–34 °C and precipitation averaging 132 cm year
. The overall site is 12,000 ha in area, 7500 ha of which are forested [
123]. The site also exists within a tributary drainage basin that eventually empties into the Flint River. Soils here are underlain by karst Ocala limestone and mostly Typic Quartzipsamments, with sporadic Grossarenic and Aquic Arenic Paleudults [
124]. Soils here often lack well-developed organic horizons [
123,
124,
125].
Forests here are mostly second-growth, approximately 65–95 years in age. Long-leaf pine (
Pinus palustris Mill.) dominates the overstory, while the understory is comprised primarily of wiregrass (
Aristida stricta Michx.) and secondarily of shrubs, legumes, forbs, immature hardwoods, and regenerating long-leaf pine forests [
126]. Prescribed fire is a regular component of management here, with stands often burned at regular 1–5 year intervals [
123]. This has promoted wiregrass and legumes in the understory, while reducing the number of hardwoods [
123]. The RD EC flux tower is contained within the mesic/intermediate sector. This site consists of only four primary tree species from two genera: Long-leaf pine (
Pinus palustris), water oak (
Quercus nigra L.), southern live oak (
Quercus virginiana Mill.), and bluejack oak (
Quercus incana W. Bartram). Measurements for the RD tower are available for the 2008–2013 time period. Summary statistics for the RD tower site for the year 2008 are provided in
Table 3.
A table of observed species abundances for the year 2009 are provided in
Table 4.
Two recent studies [
127,
128] indicate that the mesic sector of this subtropical pine savanna functions as a moderate carbon sink (NEE = −0.83 Mg C ha
year
; −1.17 Mg C ha
year
), reduced to near-neutral uptake during the 2011 drought (NEE = −0.17 Mg C ha
year
), and is a carbon source when prescribed burning is taken into account. NEE typically recovered to pre-fire rates within 30–60 days. The mechanisms behind soil respiration rates here again appear to be complex, site-specific, and poorly understood [
128].
Overall, existing research highlights the importance of fire and drought to carbon exchange in long-leaf pine (
Pinus palustris) and oak (
Quercus spp.) savanna systems [
127,
128,
129] at JERC. This is in contrast to the secondary growth-dominated deciduous broadleaf characteristics of Harvard Forest. Species diversity at the EMS tower site is 350% greater than that of the JERC-RD site, with 14 species from a variety of genera compared to four species from only two genera,
Pinus and
Quercus.
2.3. Site Data
Data collection methods may be accessed through the below data provider websites. Both sites provided a metadata file along with each data file, as is typically available to data users for the two sites. To conduct this model intercomparison exercise at HF-EMS, we leveraged the large amount of data openly available to the public through the Harvard Forest Data Archive:
Data were collected here for a range of studies, as evidenced by the Harvard Forest Data Archive. Datasets used in model validation include HF001-04, HF004-02, HF069-09, HF278-04, HF069-06, HF015-05, HF006-01, and HF069-13. These include weather station and forest inventory time-series, eddy covariance flux tower measurements, soil respiration, soil organic matter, and studies on C:N stoichiometry. Standard measurement techniques were used for each. For both sites, local tree species, age, depth-at-breast-height (DBH), biomass, soil, and meteorological data were primarily used to parameterize the models.
The Jones Ecological Research Center has hosted multiple research efforts over the years, collectively resulting in the collection of a large data library. However, JERC-RD site data are not made openly available to the public and are thus only available by request. One may find contact information located within their website:
Datasets used in model validation at JERC-RD include JC010-02, JC010-01, JC003-04, JC004-01, JC003-07, and JC011-01. These include weather station and eddy covariance flux tower measurements, forest inventory data, soil respiration, soil organic matter, and studies on C:N stoichiometry. Standard measurement techniques were also used for each of these.
2.4. Scales, Metrics, and Units
The selection of simulation years was based on the availability of EC flux tower data used in model validation. Thus, we simulated the HF-EMS site for the years 2002–2012 and the JERC-RD site for the years 2009–2013. For both sites and models, we initialized the model state in the first year of simulations using field observations. The PPA-SiBGC model used an annual timestep while LANDIS-II NECN used a monthly timestep internally. Both models may be set to other timesteps if desired.
The areal extent of the single-site model intercomparisons were designed to correspond to available field measurements. At both sites, tree inventories were conducted in 10,000
, or one-hectare, areas. All target metrics were converted to an annual areal basis to ease interpretation, comparison, and transferability of results. Importantly, an areal conversion will allow comparison to other sites around the world. While flux tower measurements for both sites were already provided on an areal (
) basis, many other variables were converted to harmonize metrics between models and study sites. For example, moles
measurements were converted to moles C through well-described molecular weights, all other measures of mass were converted to kg, and all areal and flux measurements were harmonized to
. A table of metrics and units used in the intercomparison of LANDIS-II and PPA-SiBGC is provided in
Table 5.
In the subsequent section, we describe the model intercomparison methodology.
2.5. Model Intercomparison
Intercomparison of the PPA-SiBGC and LANDIS-II models at the HF-EMS and JERC-RD EC flux tower sites was conducted using a collection of object-oriented functional programming scripts written in the R language for statistical computing [
83]. These scripts were designed to simplify model configuration, parameterization, operation, calibration/validation, plotting, and error calculation. The scripts and our parameters are available on GitHub (
https://github.com/adam-erickson/ecosystem-model-comparison), making our results fully and efficiently reproducible. The directory structure of the repository is shown in
Figure S1 in the
Supplementary Materials. The R scripts are also designed to automatically load and parse the results from previous model simulations, in order to avoid reproducibility issues stemming from model stochasticity. We use standard regression metrics applied to the time-series of observation and simulation data to assess model fitness. The metrics used include the coefficient of determination (
), root mean squared error (RMSE), mean absolute error (MAE), and mean error (ME) or bias, calculated using simulated and observed values. Our implementation of
follows the Bravais–Pearson interpretation as the squared correlation coefficient between observed and predicted values [
130]. This implementation is provided in Equation (
2).
where
n is the sample size,
is the
ith observed value,
is the
ith predicted value,
is the mean observed value, and
is the mean predicted value. The calculation of RMSE follows the standard formulation, as shown in Equation (
3).
where
n is the sample size and
is the error for the
tth value, or the difference between observed and predicted values. The calculation of MAE is similarly unexceptional, per Equation (
4).
where again
n is the sample size and
is the error for the
tth value. Our calculation of mean error (ME) or bias is the same as MAE, but without taking the absolute value.
While Nash–Sutcliffe efficiency (NSE) is often used in a simulation model context, we selected the Bravais–Pearson interpretation of
over NSE to simplify the interpretation of results. The NSE metric replaces
with
, where
is the sum of squares. Thus, NSE is analogous to the standard
coefficient of determination used in regression analysis [
131]. The implementation of
that we selected is important to note, as its results are purely correlative and quantify only dispersion, ranging in value between zero and one. This has some desirable properties in that no negative or large values are produced, and that it is insensitive to differences in scale. Regardless of the correlation metric used, complementary metrics are needed to quantify the direction (i.e., bias) and/or magnitude of error. We rely on RMSE and MAE to provide information on error or residual magnitude, and ME to provide information on bias. We utilize a visual analysis to assess error directionality over time, as this can be poorly characterized by a single coefficient, masking periodicity.
We compute
, RMSE, MAE, and ME for time-series of the metrics described in
Table 5 on page 13. These include NEE, above- and below-ground biomass, C, and N, soil organic C and N, soil respiration (
), aboveground net primary production (ANPP), and, species aboveground biomass and relative abundance. All of these metrics are pools with the exception of NEE,
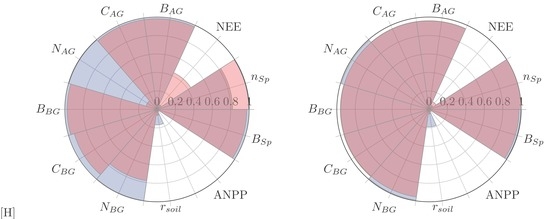
, and ANPP fluxes. Finally, we diagnose the ability of both models to meet a range of logistical criteria related to deployment: Model usability, performance, and transferability. Model usability is assessed per four criteria:
Model software performance is assessed per a single metric: The speed of program execution for each site for the predefined simulation duration. The durations are 11 years and five years for the HF-EMS and JERC-RD EC flux tower sites, respectively. Simulation results are output at annual temporal resolution, the standard resolution for both models, while NECN operates on a monthly timestep, and most other modules of LANDIS-II are annual. Finally, model transferability is assessed per the following five criteria:
Each of these logistical criteria are compared in a qualitative analysis, with the exception of software performance.
3. Results
Both PPA-SiBGC and LANDIS-II NECN showed strong performance for pools at the two model intercomparison sites, frequently achieving
values approaching unity. Yet, both models showed weak performance for fluxes. The models failed to accurately predict ANPP, while PPA-SiBGC showed stronger
performance and LANDIS-II NECN showed stronger NEE performance. The
values for both models and sites are visualized in
Figure 5.
On average, PPA-SiBGC outperformed LANDIS-II NECN across the sites and metrics tested, showing higher correlations, lower error, and less bias overall (HF-EMS , , ; JERC-RD , , ). This result is based on calculating mean values for , RMSE, MAE, and ME in order to clearly translate the overall results. The two models produced the following mean values for each of the four statistical metrics and two sites:
As shown in
Table 6, PPA-SiBGC yielded higher
values and lower RMSE, MAE, and ME values in comparison to LANDIS-II, on average, across all sites and metrics tested. Below, we provide model intercomparison results individually for the two sites, HF-EMS and JERC-RD.
3.1. HF-EMS
For the HF-EMS site, PPA-SiBGC showed higher
values and lower RMSE, MAE, and ME values compared to LANDIS-II NECN across the range of metrics. While PPA-SiBGC predicted NEE and species relative abundance showed weaker correlations with observed values compared to LANDIS-II NECN, the magnitude of error was lower, as evidenced by lower RMSE, MAE, and ME values. While LANDIS-II NECN showed a lower magnitude of error for belowground N, this is the only metric where this is the case, while the correlation of this metric to observed values was also lower than that of PPA-SiBGC. Overall results for the HF-EMS site model intercomparison are shown in
Table 7.
Time-series figures allow a visual analysis of the temporal dynamics between observations and model predictions in order to assess the ability of models to capture interannual variability in carbon exchange. Both models effectively captured integrals of dynamics in biomass, C, and, species biomass and abundance. In
Figure 6, the temporal differences in modeled NEE, aboveground C, ANPP, and soil respiration are shown for the two models in comparison to observations for the HF-EMS site. LANDIS-II NECN predicted NEE showed a higher correlation with observations while the magnitude of error and bias were lower. Furthermore, LANDIS-II NECN predicted that the HF-EMS site is a net C source, rather than sink, in contrary to observations. Meanwhile, PPA-SiBGC outperformed LANDIS-II NECN in aboveground C per both
and RMSE. Both models overpredicted species cohort biomass, while LANDIS-II NECN underpredicted total aboveground C.
An analysis of simulated species biomass and abundance also shows greater fidelity of the PPA-SiBGC model to data, as shown in
Figure 7. As LANDIS-II NECN does not contain data on individual trees, species relative abundance is calculated based on the number of cohorts of each species. Two species were simulated in LANDIS-II NECN, as there are no explicit trees in the model and the number of cohorts appears to have no effect on the total biomass. Results for PPA-SiBGC indicate that species relative abundance may be improved in future studies by optimizing mortality and fecundity rates. Meanwhile, species biomass predictions output by LANDIS-II NECN were inverted from those of the observations.
3.2. JERC-RD
For the JERC-RD site, both models showed stronger fidelity to data than for the HF-EMS site. Again, PPA-SiBGC showed higher
values and lower RMSE and MAE values compared to LANDIS-II NECN across the range of metrics tested. Yet, the margin between models was smaller for the JERC RD site. While PPA-SiBGC demonstrated higher correlations and lower errors for most metrics tested, LANDIS-II NECN outperformed PPA-SiBGC in a few cases. This includes a higher correlation for NEE, ANPP, and lower magnitude of error for aboveground N, belowground biomass, soil respiration, and SOC. PPA-SiBGC, however, showed correlations equal or higher for all metrics tested, and lower errors for all other metrics. Overall results for the JERC-RD site model intercomparison are shown in
Table 8.
Time-series carbon exchange metrics for the JERC-RD site, presented in
Figure 8, show that modeled NEE values are positively correlated with each other rather than with observed NEE, while the magnitude of error varies from favoring PPA-SiBGC to LANDIS-II NECN. Overall, the PPA-SiBGC model shows a lower magnitude of error for NEE, ANPP, and
, and slightly higher for
. Again, for
the two models show strong agreement, but underestimate observations by an order of magnitude. For
and ANPP, PPA-SiBGC shows good overall fit.
While both models showed higher performance at the JERC-RD site in comparison to the HF-EMS site, an analysis of simulated species biomass and abundance again indicates greater fidelity of the PPA-SiBGC model to data, as shown in
Figure 9. While LANDIS-II NECN greatly overpredicts the rate of longleaf pine growth, PPA-SiBGC matches observed species abundance and biomass trajectories for all species present. While the correlations are high, PPA-SiBGC overpredicts the magnitude of species biomass.
Our results for the HF-EMS and JERC-RD site model intercomparison exercise show strong performance for both models at both sites. Results for the JERC-RD site are particularly close between the two models. Next, we assess results related to the logistics of model deployment to new computers, users, and modeling sites.
3.3. Model Usability, Performance, and Transferability
While the two models share a similar basis in forest dynamics and biogeochemistry modeling, they differ in important practical and conceptual terms. The command-line version of the PPA-SiBGC model used in this work, version 5.0, consists of approximately 500 lines of R code and is thus readily cross-platformed and portable. Meanwhile, the LANDIS-II model core and NECN succession extension are an estimated 2000 and 0.5 million lines of code, respectively. While this version of PPA-SiBGC fuses an explicit tree canopy geometry model with empirical data on fecundity, growth, mortality, and stoichiometry, the NECN extension of LANDIS-II borrows heavily from the process-based Century model [
37], similar to the MAPSS-Century-1 (MC1) model [
132]. This carries important implications for model parameterization needs. While PPA-SiBGC relies on typical forest inventory data, including tree species, age/size, and densities, LANDIS-II relies on species age/size and traits in the form of vital attributes, in addition to approximately 100 NECN parameters. Below, we summarize our findings regarding the logistics of model deployment.
3.3.1. Model Usability
In the following section, we provide an assessment of model usability based on four criteria.
Ease of installation
While LANDIS-II NECN requires the installation of two Windows programs, depending on the options desired, PPA-SiBGC is contained in a single R script and requires only a working R installation.
Ease of parameterization
While both models can be difficult to parameterize for regions with little to no observational data, the simple biogeochemistry in PPA-SiBGC requires an order of magnitude fewer parameters than LANDIS-II NECN. In addition, PPA-SiBGC uses commonly available forest inventory data while NECN requires a number of parameters that may be difficult to locate.
Ease of program operation
Both models use a command-line interface and are thus equally easy to operate. Yet, PPA-SiBGC is cross-platform and uses comma-separated-value (CSV) files for input tables, which are easier to work with than multiple tables nested within an unstructured text files. This additionally allows for simplification in designing model application programming interfaces (APIs), or model wrappers, a layer of abstraction above the models. These abstractions are important for simplifying model operation and reproducibility, and enable a number of research applications.
Ease of parsing outputs
All PPA-SiBGC outputs are provided in CSV files in a single folder while LANDIS-II NECN generates outputs in multiple formats in multiple folders. While the PPA-SiBGC format is simpler and easier to parse, the image output formats used by LANDIS-II carry considerable benefit for spatial applications. Both models may benefit by transitioning spatiotemporal data to the NetCDF scientific file format used by most general circulation and terrestrial biosphere models.
3.3.2. Model Performance
Next, we assess model performance in terms of the speed of operation on a consumer-off-the-shelf (COTS) laptop computer with a dual-core 2.8 GHz Intel Core i7-7600U CPU and 16 GB of DDR4-2400 RAM. We focus on a single performance metric, the timing of simulations. Other aspects of model performance in the form of precision and accuracy are described in previous sections. As shown in
Table 9, PPA-SiBGC was between 1200 and 2800% faster than LANDIS-II NECN in our timing tests. This was surprising given that PPA-SiBGC models true cohorts (i.e., individual trees) in an interpreted language while LANDIS-II models theoretical cohorts (i.e., cohorts without a physical basis) in a compiled language. The difference in speed is likely attributable to the parsimony of the PPA-SiBGC model.
3.3.3. Model Transferability
Here, we discuss model transferability. In this section, we assess the effort required to transfer the models to new locations, new computer systems, or new users. All three are important logistical criteria for effective model deployment.
Model generalization
Both models appear to generalize effectively to different forested regions of the world, as both have shown strong performance in this study and others. No clear winner is evident in this regard. In terms of model realism, PPA-SiBGC has a more realistic representation of forest canopies while LANDIS-II NECN has more realistic processes, as it is a Century model variant.
Availability of parameterization data
While LANDIS-II NECN requires substantially greater parameterization data compared to PPA-SiBGC, it may often be possible to rely on previously published parameters. Meanwhile, the growth, mortality, and fecundity parameters used by PPA-SiBGC are easy to calculate using common field inventory data. PPA-SiBGC is simpler to transfer in this regard given the wide availability of forest inventory data.
Size of the program
PPA-SiBGC is approximately 500 lines of R code, while LANDIS-II NECN is estimated at 0.5 million lines of C# code.
Cross-platform support
While Linux support may soon be supported with Microsoft .NET Core, LANDIS-II NECN is written in C# and is thus limited to Microsoft Windows platforms. Meanwhile, PPA-SiBGC is written in standard R code and is fully cross-platform.
Ease of training new users
While both models have a learning curve, the practical simplicity of PPA-SiBGC may make it easier to train new users. While LANDIS-II NECN contains more mechanistic processes and related parameters, these come at the cost of confusing new users. The model wrapper library we developed as part of this work vastly eases the operation of both models. Future studies should measure the time required for new users to effectively operate both models.
4. Discussion
First, it is important to clarify some terms used in this analysis. Gross primary production (GPP) is the net rate of carboxylation and oxygenation by RuBisCO and is calculated as
, where
is gross photosynthesis and
is photorespiration. In EC flux data analyses, GPP is also known as gross ecosystem exchange (GEE) or gross ecosystem production (GEP) and is often estimated inversely from NEE or NEP flux tower retrievals as
, where
is ecosystem respiration or the sum of auto- and heterotrophic respiration components. Thus,
where
is maintenance respiration,
is autotrophic growth respiration, and
is heterotrophic respiration. While GPP is the total amount of C fixed by plants in photosynthesis, NPP subtracts autotrophic respiration (
) as
where
. NEE or net ecosystem production (NEP) is then calculated as NPP minus heterotrophic respiration, or
, which is equivalent to
. During the day,
while during the night,
and
are absent, making NEE approximate to ecosystem respiration, or
. Traditionally, gross or net exchange of
into the forest is negative and fluxes into the atmosphere are positive, while each constituent process is discussed with a positive sign. Thus, NEE is often calculated as
where each constituent flux term is always positive [
133,
134,
135,
136,
137].
All this is to say that there exists much difficulty in relating NPP from field inventories and soil respiration samples directly to NEE from EC flux towers, integrated over the year. In our analyses, we assume that the observed annual biomass growth increment is equivalent to ANPP and that soil respiration (
) is equivalent to ecosystem respiration (
), or
. Yet, there are known error contributions at multiple conversion points, making the comparison of models based on field data and EC flux tower measurements difficult. For example, field inventory estimates of ANPP contain known sources of error in converting DBH to biomass, both above- and belowground [
115], and there are additional errors in converting biomass to C based on a fixed fraction for each biomass compartment. Meanwhile, unlike
,
does not account for
or
, only
. Even if these fluxes were approximately similar, spatial biases in the EC flux tower footprint or contributing area [
138,
139,
140,
141,
142] may make field inventory and tower measurements difficult to harmonize.
As others have noted [
118,
119], including a previous study on flux measurements at the HF-EMS site [
88], it is evident that treating ANPP as the C fraction of woody biomass increment per allometric relations from field data is a loose proxy for ecosystem ANPP, given its visible disconnection from observed NEE and
fluxes. Given the definition of NEE, the relation between these variables should be approximately linear. While others have reported hysteresis between peaks in NEE and growth increment at the HF-EMS site [
88], we did not see evidence of this dynamic. Instead, flux tower NEE appears to have little to no connection to field data ANPP and observed
fluxes at both sites in this analysis. Nevertheless, both models showed good agreement with net changes to C and N pools. This may partially reflect difficulties in accounting for belowground processes, which can contribute disproportionately to C fluxes, and in connecting flux tower NEE to forest stands where the contributing area extent is far greater than a one-hectare stand, as is often the case [
138,
139,
140].
This issue can be seen in
Figure 6 and
Figure 8. In this model intercomparison exercise, ANPP for the PPA-SiBGC model and field data are based on annual woody biomass increment, while ANPP in LANDIS-II NECN includes the Century process model for estimating ANPP. Rather than this basis making the NECN model purely process-based or mechanistic, species-specific growth is tightly constrained by empirical limits in a truncated logistic curve, with LAI and the number of cohorts present used as a proxy for growing space limitations and moisture and temperature used for physiological constraint based on Liebig’s Law of the Minimum. In contrast, PPA-SiBGC is parameterized with mean observed growth and mortality rates from field data, which vary depending on the canopy position of a cohort. Understory cohorts assumed to be in full shade face higher mortality and lower growth, as is widely evident in field data, while overstory cohorts assumed to be in full sunlight have higher growth and lower mortality rates. While soil and root processes are explicitly simulated in Century and thus LANDIS-II NECN, PPA-SiBGC relies on simple stoichiometric and allometric relations from field data to model these pools. In other words, PPA-SiBGC is designed primarily to model pools rater than fluxes, as the former are of generally higher interest to foresters.
The strong empirical basis of parameterization of both PPA-SiBGC and LANDIS-II NECN explains why the two models are often in better agreement with each other than with observations. The similarity of outputs from the two models is perhaps surprising, given their differences in model architecture and theoretical basis. This shows that, despite any mechanistic process present, both models in their current form are closely fit to field data and are therefore strongly empirical, as evidenced by their representation of growth processes. Meanwhile, this design choice limits the representation of fluxes in both models, as detailed process models are absent. This is expected for PPA-SiBGC, which is intended primarily to be a simple empirical pool model. This work also shows that observations between field and tower measurements are substantially disconnected. We estimate that fluxes are poorly represented by both models because they are tightly coupled to field inventory data rather than to tower-based measurements. Hence, patterns evident in field inventory data are reliably reproduced while fluxes appear wholly uncoupled.
The advancement of processor architectures has facilitated the development of increasingly complex forest models. Each new generation of processors allows researchers to conduct large-scale simulations faster and more efficiently than previous designs. As a result, forest models have grown into large, complex, analytically intractable programs. Rigorous intercomparison of models developed by different research groups, as well as the diagnosis of new versions of established models, is therefore a critical step in further advancing ecosystem models. This ensures that models are properly diagnosed and compared in a consistent, reliable, and transparent manner. Too often, model intercomparisons are conducted by each separate research group applying their own model in a manner that is, at best, inconsistent and opaque. In this work, we extended our model intercomparison by further providing wrapper functions that may be used to benchmark additional models or sites through a unified modeling framework. This ensures the consistency and transparency of intercomparison results.
The presented research is intended to establish the groundwork for future model intercomparison studies at both sites in order to advance the design of new models. Furthermore, we hope that this work will inspire a new generation of forest model intercomparisons in North America, which are sorely absent. Forest models have proven to be a critical testbed for improving the representation of vegetation dynamics in global terrestrial biosphere models [
40,
41,
143], given the importance of forests in the global carbon cycle and the increased detail of local- to regional-scale models. Model benchmarking datasets and related results should be publicly shared and regularly updated with version-controlled software repositories (e.g., GitHub or GitLab), as is commonplace in the machine learning research community. Cloud computing providers may provide full reproducibility for cases where compute is limiting. In general, there is a broad disparity between modern software tools and existing forest models.
One important new forest model in development is a next-generation model from the SORTIE-PPA family of models, known as SORTIE-NG. This new model combines mechanistic representations of demographic processes, energetic and biogeochemical fluxes, and landscape disturbance dynamics, using hierarchical multiscale modeling with a modular component-based software framework [
144]. Along with LM3-PPA [
45], SORTIE-NG is among the first of a new class of hybrid models that we term ‘cohort-leaf’ models for their partitioning of energetic and biogeochemical fluxes amongst dynamic vegetation cohorts, instead of a single vertical ’big-leaf’ profile. The SORTIE-NG model includes evolutionary optimality principles as well as phenotype plasticity and intraspecific genetic diversity through first-class support for probabilistic modeling, borrowing design principles from probabilistic programming languages (e.g., [
145]). Thus, SORTIE-NG is intended to be the first forest model to bridge the divide between big-leaf, gap, and landscape models, and to be designed from the outset as a probabilistic modeling framework [
144]. Future model extensions are in the planning stages, including the first machine learning processes included in an ecosystem model.
While implemented in a ’close-to-metal’ language (i.e., C++17) and designed for efficiency, SORTIE-NG is more computationally demanding than the PPA-SiBGC model used in this paper. Yet, we anticipate that SORTIE-NG will be able to improved the fidelity to observed fluxes through reliance on detailed process models, which is the major shortcoming of both models considered in this paper. Similarly, there is a new version of the LANDIS-II NECN model in development known as NECN-Hydro, which remains a simplified variant of the Century model, but includes more detailed hydrological processes. The currently presented work provides not only an intercomparison of two current state-of-art models, but also open-source software and wrapper functions for simple and rapid comparison of our results with new models or sites. The selected forested ecosystems modeled in this work are among the best-studied model forests on Earth today. Specifically, the EMS EC flux tower at Harvard Forest is the longest running flux tower in the United States. Extensions of the presented work will allow rigorous model comparison methodologies for forest models that will benefit the research community at large.
Extensions of this work may also address the robustness of model predictions to variations in parameter values. The parameterization of complex forest biogeochemistry models such as LANDIS-II NECN and PPA-SiBGC is an important problem for consideration. Models such as LANDIS-II NECN operate with an order of magnitude more parameters than PPA-SiBGC, which can each be estimated with different levels of accuracy. Often, we know only the range of parameter values while parameterization can also depend on the statistical approach employed. Meanwhile, authors routinely employ additional model calibration that consists of adjusting parameters in order to obtain improved fitness, which we explicitly avoided in this study.
Conducting such analyses through a unified software framework in a fully transparent and reproducible manner is therefore of the utmost importance. This is exactly the type of analyses that our provided software is designed to support. In a parallel line of research, we extend this base-level implementation into a generic application programming interface (API) and toolkit for geoscientific simulation models, known as Erde [
146], supporting both R and Python. The Erde framework provides machine learning model emulation, robust loss estimation, parameter optimization, probabilistic parameterization, samplers such as Latin hypercube sampling and Markov Chain Monte Carlo, and a number of other helper methods designed for complex simulation models. We utilize the Erde framework in the design of Erde Gym, a toolkit for developing and comparing optimization algorithms in the geosciences with a focus on reinforcement learning [
146]. For the first time, Erde Gym will allow us to model systems (e.g., evolutionary plant optimality) as intelligent agents able to navigate complex environments.
4.1. Limitations
This study, similar to most other modeling studies, was limited by the availability, quality, and quantity of observational data. The lack of temporal depth in this data poses substantial challenges in modeling the long-term effects of forest succession, as these processes can operate on a century timescale or longer. However, diagnosing succession was not the aim of this study, as we instead focus on near-term validation of forest models using field measurements and EC flux tower data. Another limitation is that these methods may be challenging to implement for sites that are less well-characterized, particularly in the absence of EC flux tower data and/or tree species parameters. A combination of tower-based and remote sensing observations may help overcome this challenge in the coming years with advances in machine learning. In addition, the poor performance of both tested models in capturing fluxes and excellent performance in capturing stocks indicate that the two current models should be applied in cases where stocks, rather than fluxes, are of primary interest.
4.2. Future Opportunities
Future studies should expand upon the PPA with a first-principles representation of energetic and biogeochemical above- and below-ground processes in a modern component-based software framework. This work should fuse the new state-of-the-art forest biogeochemistry model with a model wrapper API written in R or Python, in order to expand native model functions to include Monte Carlo methods, machine-learning model emulation, robust loss functions, and optimization through a simple API enabling reproducibility. This would combine a high-performance forest model written in a compiled language with a simple, user-friendly interface written in an interpreted language, combining the best of both worlds. We are currently conducting work along this line by fusing the SORTIE-NG model with the Erde framework in order to develop state-of-the-art and user-friendly modeling capabilities, inspired by the design of modern deep learning frameworks such as PyTorch [
147] and the Keras API [
148].
In addition, there is a clear opportunity to link individual-based models such as PPA-SiBGC and SORTIE-NG to remote sensing data including airborne laser scanning or high-resolution multiview-stereo imagery (i.e., structure-from-motion), and hyperspectral indices of vegetation growth or stress. This line of work may assess opportunities for Bayesian data assimilation in addition to model parameterization and validation using detailed wall-to-wall forest structure maps. As models such as LES [
32] provide more structural detail, spatially explicit data will be needed to parameterize the next generation of models. New data collection methods (e.g., [
149]) will also be needed as the geometric realism of models advances toward the photorealistic detail offered by procedural models such as Lindenmayer- or L-systems [
150,
151].