Synthetic (E)-3-Phenyl-5-(phenylamino)-2-styryl-1,3,4-thiadiazol-3-ium Chloride Derivatives as Promising Chemotherapy Agents on Cell Lines Infected with HTLV-1
Abstract
:1. Introduction
2. Results and Discussion
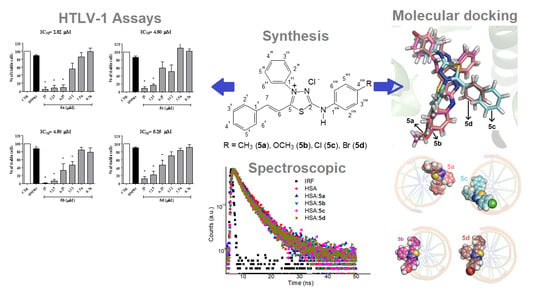
2.1. Synthesis
2.2. Biological Activity
2.3. Interaction with HSA
2.3.1. Experimental Binding Ability
2.3.2. Perturbation on the Surface and Secondary Structure of Albumin
2.4. Molecular Docking Evaluation
3. Materials and Methods
3.1. General Information
3.2. General Procedure for the Synthesis of Aryl-thioureas (2a–2d)
3.3. General Procedure for the Synthesis of Aryl-isothiocyanates (3a–d)
3.4. General Procedure for the Preparation of Hydrazinecarbothiamides (4a–d)
3.5. General Procedure for the Preparation of Mesoionic Chlorides (5a–d)
3.6. Biological Assays
3.6.1. Cell Viability Assays
3.6.2. Reactive Oxygen Species (ROS) Detection
3.6.3. DNA Interaction Assay
3.6.4. Statistical Analysis
3.7. Spectroscopic HSA Binding Studies
3.8. Zeta Potential Measurements for HSA Surface Perturbation
3.9. Molecular Docking Procedure for DNA and HSA
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [PubMed] [Green Version]
- Pui, C.H.; Robison, L.L.; Look, A.T. Acute lymphoblastic leukaemia. Lancet 2008, 371, 1030–1043. [Google Scholar] [PubMed]
- Uchiyama, T.; Yodai, J.; Sagawa, K.; Takatsuki, K.; Uchino, H. Adult T-cell leukemia: Clinical and hematologic features of 16 cases. Blood 1977, 50, 481–492. [Google Scholar] [PubMed]
- Gessain, A.; Barin, F.; Vernant, J.C.; Gout, O.; Maurs, L.; Calender, A.; de Thé, G. Antibodies to human T-lymphotropic virus type-I in patients with tropical spastic paraparesis. Lancet 1985, 326, 407–510. [Google Scholar]
- Poiesz, B.J.; Ruscetti, F.W.; Gazdar, A.F.; Bunn, P.A.; Mina, J.D.; Gallo, R.C. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc. Natl. Acad. Sci. USA 1980, 77, 7415–7419. [Google Scholar] [CrossRef] [Green Version]
- Gessain, A.; Cassar, O. Epidemiological aspects and world distribution of HTLV-1 infection. Front. Microbiol. 2012, 3, 388–411. [Google Scholar] [CrossRef] [Green Version]
- Iwanaga, M.; Watanabe, T.; Yamaguchi, K. Adult T-cell leukemia: A review of epidemiological evidence. Front. Microbiol. 2012, 3, 322–335. [Google Scholar]
- Prince, H.E.; York, J.; Golding, J.; Owen, S.M.; Lal, R.B. Spontaneous lymphocyte proliferation in human T-cell lymphotropic virus type-I (HLTV-I) and HTLV-II infection: T-cell subset responses and their relationships to the presence of provirus and viral antigen production. Clin. Diagn. Lab. Immunol. 1994, 1, 237–282. [Google Scholar] [CrossRef]
- Boxus, M.; Willems, L. Mechanisms of HTLV-I persistence and transformation. Br. J. Cancer 2009, 101, 1497–1501. [Google Scholar] [CrossRef] [Green Version]
- Shimoyama, M. Diagnostic criteria and classification and clinical subtypes of adult T-cell leukemia-lymphoma. A report from the Lymphoma Study Group. Br. J. Haematol. 1991, 79, 428–437. [Google Scholar] [CrossRef]
- Matsuoka, M. Human T-cell leukemia virus type I and adult T-cell leukemia. Oncogene 2003, 22, 5131–5140. [Google Scholar]
- Ishitsuka, K.; Tamura, K. Human T-cell leukaemia virus type I and adult T-cell leukaemia-lymphoma. Lancet Oncol. 2014, 15, 517–526. [Google Scholar]
- Mehta-Shah, N.; Ratner, L.; Horwitz, S.M. Adult T-cell leukemia/lymphoma. J. Oncol. Pract. 2017, 13, 487–489. [Google Scholar] [CrossRef]
- Tsukasaki, K.; Hermine, O.; Bazarbachi, A.; Ratner, L.; Ramos, J.C.; Harrington, W., Jr.; O’Mahony, D.; Janik, J.E.; Bittencourt, A.L.; Taylor, G.P.; et al. Definition, prognostic factors, treatment and response criteria of adult T-cell leukemia-lymphoma: A proposal from an international consensus meeting. J. Clin. Oncol. 2009, 27, 453–459. [Google Scholar]
- Yared, J.A.; Kimball, A.S. Optimizing management of patients with adult T cell leukemia-lymphoma. Cancers 2015, 7, 2318–2329. [Google Scholar] [CrossRef]
- Marino-Merlo, F.; Mastino, A.; Grelli, S.; Hermine, O.; Bazarbachi, O.; Macchi, B. Future perspectives on drug targeting in adult T cell leukemia-lymphoma. Front. Microbiol. 2018, 9, 925–933. [Google Scholar] [CrossRef]
- Hermine, O.; Ramos, J.C.; Tobinai, K. A review of new findings in adult T-cell leukemia-lymphoma: A focus on current and emerging treatment strategies. Adv. Ther. 2018, 35, 135–152. [Google Scholar] [CrossRef] [Green Version]
- Phillips, A.A.; Harewood, J.C.K. Adult T cell Leukemia-lymphoma (ATL): State of the art. Curr. Hematol. Malig. Rep. 2018, 13, 300–307. [Google Scholar] [CrossRef]
- Ishida, T.; Takemoto, S.; Suzushima, H.; Uozumi, K.; Yamamoto, K.; Uike, N.; Saburi, Y.; Nosaka, K.; Utsunomiya, A.; Tobinai, K.; et al. Dose-intensified chemotherapy alone or in combination with mogamulizumab in newly diagnosed aggressive adult T-cell leukaemia-lymphoma: A randomized phase II study. Br. J. Haematol. 2015, 101, 398–404. [Google Scholar] [CrossRef]
- Cascioferro, S.; Petri, G.L.; Parrino, B.; El Hassouni, B.; Carbone, D.; Arizza, V.; Perricone, U.; Padova, A.; Funel, N.; Peters, G.J.; et al. 3-(6-phenylimidazo [2,1-b][1,3,4-thiadiazol-2-yl]-1H-indole derivatives as new anticancer agents in the treatment of pancreatic ductal adenocarcinoma. Molecules 2020, 25, 329. [Google Scholar] [CrossRef] [Green Version]
- Faraooqi, S.I.; Arshad, N.; Channar, P.A.; Perveen, F.; Saeed, A.; Larik, F.A.; Javeed, A. Synthesis, theoretical, spectroscopic and electrochemical DNA binding investigations of 1,3,4-thiadiazole derivatives of ibuprofen and ciprofloxacin: Cancer line studies. J. Photochem. Photobiol. B 2018, 189, 104–118. [Google Scholar] [CrossRef]
- Brandt, A.P.; Gozzi, G.J.; Pires, A.R.A.; Martinez, G.R.; Canuto, A.V.S.; Echevarria, A.; Di Pietro, A.; Cadena, S.M.S.C. Impairment oxidative phosphorylation increase the toxicity of SYD-1 on hepatocarcinoma cells (HepG2). Chem. Biol. Interact. 2016, 256, 154–160. [Google Scholar] [CrossRef]
- Dos Reis, C.M.; Echevarria-Lima, J.; Miranda, A.F.; Echevarria, A. Improved synthesis of 1,3,4-thiadiazolium-2-phenylamines using microwave and ultrasound irradiation and investigation of their cytotoxic activity. J. Braz. Chem. Soc. 2011, 22, 1505–1510. [Google Scholar] [CrossRef] [Green Version]
- Soares-Bezerra, R.J.; Leon, L.L.; Echevarria, A.; Reis, C.M.; Gomes-Silva, L.; Agostinho, C.G.; Fernandes, R.A.; Canto-Cavalheiro, M.M.; Genestra, M. In vitro evaluation of 4-phenyl-5-(4′-X-phenyl)-1,3,4-thiadiazolium-2-phenylaminide chlorides and 3-[N-4′-X-phenyl-1,2,3-oxadiazolium-5-olate derivatives on nitric oxide synthase and arginase activity of Leishmania amazonensis. Exp. Parasitol. 2013, 135, 50–54. [Google Scholar] [CrossRef] [Green Version]
- Rodrigues, R.F.; Charret, K.S.; Campos, M.C.; Amaral, V.; Echevarria, A.; Reis, C.M.; Canto-Cavalheiro, M.M.; Leon, L.L. The in vivo activity of 1,3,4-thiadiazolim-2-aminide compounds in the treatment of cutaneous and visceral leishmaniasis. J. Antimicrob. Chemother. 2012, 65, 182–190. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, W.S.; Freire-de-Lima, L.; Saraiva, F.; Alisson-Silva, V.B.; Mendonça-Previato, L.; Previato, J.O.; Echevarria, A.; Lima, M.E.F. Novel 1,3,4-thiadiazolium-2-phenylamine chlorides as trypanocidal agents: Chemical and biological studies. Bioorg. Med. Chem. 2008, 16, 2984–2991. [Google Scholar] [CrossRef]
- Kalluraya, B.; Aamir, S.; Shabaraya, A.R. Regioselective reaction: Synthesis, characterization and pharmacological activity of some new Mannich and Schiff bases containing sydnone. Eur. J. Med. Chem. 2012, 54, 597–604. [Google Scholar] [CrossRef]
- Ragab, F.A.; Heiba, H.I.; El-Gazzar, M.G.; Abou-Seri, S.M.; El-Sabaagh, W.A.; El-Hazek, R.M. Anti-inflammatory, analgesic and COX-2 inhibitory activity of novel thiadiazoles in irradiated rats. Photochem. Photobiol. 2017, 166, 285–300. [Google Scholar] [CrossRef]
- Paiva, R.O.; Kneipp, L.F.; Reis, C.M.; Echevarria, A. Mesoionic compounds with antifungal activity against Fusarium verticillioides. BMC Microbiol. 2015, 15, 11–19. [Google Scholar] [CrossRef] [Green Version]
- Abo-Bakr, A.M.; Hashem, H.E. New 1,3,4-thiadiazole derivatives: Synthesis, characterization and antimicrobial activity. J. Heterocycl. Chem. 2019, 56, 1038–1047. [Google Scholar] [CrossRef]
- Ollis, W.D.; Stanforth, S.P.; Ramsden, C.A. Heterocyclic mesomeric betaines. Tetrahedron 1985, 41, 2239–2329. [Google Scholar] [CrossRef]
- Gozzi, G.J.; Pires, A.R.A.; Valdameri, G.; Rocha, M.E.M.; Martinez, G.R.; Noleto, G.R.; Acco, A.; Souza, C.E.A.; Echevarria, A.; Reis, C.M.; et al. Selective cytotoxicity of 1,3,4-thiadiazolium mesoionic derivatives on hepatocarcinoma cells (HepG2). PLoS ONE 2015, 10, e0130046–e0130053. [Google Scholar] [CrossRef]
- Pires, A.R.A.; Noleto, G.R.; Echevarria, A.; Reis, C.M.; Rocha, M.E.M.; Carnieri, E.G.S.; Martinez, G.R.; Cadena, S.M.S.C. Interaction of 1,3,5-thiadiazolium mesoionic derivatives with mitochondrial membrane and scavenging activity: Involvement of their effects on mitochondrial energy-linked functions. Chem. Biol. Interact. 2011, 189, 17–25. [Google Scholar] [CrossRef]
- Mendez-Sanchez, S.C.; Martinez, G.R.; Romão, S.; Echevarria, A.; Silva, E.F.; Rocha, M.E.M.; Noleto, G.R.; Cadena, S.M.S.C.; Oliveira, M.B.M. The inhibition of lipoperoxidation by mesoionic compound MI-D: A relationship with uncoupling effect and scavenging activity. Chem. Biol. Interact. 2009, 179, 125–130. [Google Scholar] [CrossRef]
- Romão, S.; Cadena, S.M.S.C.; Amorim, J.C.; Mendez-Sanchez, S.C.; Silva, E.F.; Rocha, M.E.M.; Noleto, G.R.; Carnieri, E.G.S.; Martinez, G.R.; Oliveira, M.B.M. Metabolism of mesoionic compound (MI-D) by mouse liver microsome, detection of its metabolite, in vivo and acute toxicity in mice. J. Biochem. Mol. Toxicol. 2009, 23, 394–405. [Google Scholar] [CrossRef]
- Senff-Ribeiro, A.; Echevarria, A.; Silva, E.F.; Veiga, S.S.; Oliveira, M.B.M. Effect of new 1,3,4-thiadiazolium mesoionic compound (MI-D) on B16-F10 murine melanoma. Melanoma Res. 2003, 13, 465–471. [Google Scholar] [CrossRef]
- Senff-Ribeiro, A.; Echevarria, A.; Silva, E.F.; Veiga, S.S.; Oliveira, M.B.M. Antimelanoma activity of 1,3,4-thiadiazolium mesoionics: A structure-activity relationship study. Anti-Cancer Drugs 2004, 15, 269–275. [Google Scholar] [CrossRef]
- Xiaohe, Z.; Yu, Q.; Hong, Y.; Xiuqing, S.; Rugang, Z. Synthesis, biological evaluation and molecular modeling studies of N-aryl-2-arylthioacetamides as non-nucleoside HIV-1 reverse transcriptase inhibitors. Chem. Biol. Drug Des. 2010, 76, 330–339. [Google Scholar] [CrossRef]
- Shafique, M.; Hameed, S.; Naseer, M.M.; Al-Masoudi, N.A. Synthesis of new chiral 1,3,4-thiadiazole-based di- and tri-arylsulfonamide residues and evaluation of in vitro anti-HIV activity and cytotoxicity. Mol. Divers. 2018, 22, 957–968. [Google Scholar] [CrossRef]
- Buemi, M.R.; Gitto, R.; Ielo, L.; Pannecouque, C.; De Luca, L. Inhibition of HIV-1 RT activity by a new series of 3-(1,3,4-thiadiazol-2-yl)-thiazolidine-4-one derivatives. Bioorg. Med. Chem. 2020, 28, 115431. [Google Scholar] [CrossRef]
- Zhang, H.; Deng, H.; Wang, Y. Comprehensive investigations about the binding interaction of acesulfame with human serum albumin. Spectrochim. Acta A 2020, 237, 118410. [Google Scholar] [CrossRef] [PubMed]
- Singh, I.; Luxami, V.; Paul, K. Spectroscopy and molecular docking approach for investigation on the binding of nocodazole to human serum albumin. Spectrochim. Acta A 2020, 235, 118289. [Google Scholar]
- Almutairi, F.M.; Ajmal, M.R.; Siddiqi, M.K.; Amir, M.; Khan, R.H. Multi-spectroscopic and molecular docking technique study of the azelastine interaction with human serum albumin. J. Mol. Struct. 2020, 1201, 127147. [Google Scholar] [CrossRef]
- He, X.M.; Carter, D.C. Atomic structure and chemistry of human serum albumin. Nature 1992, 358, 209–215. [Google Scholar] [CrossRef] [Green Version]
- Wardell, M.; Wang, Z.; Ho, J.X.; Robert, J.; Rucker, F.; Ruble, J.; Carter, D.C. The atomic structure of human methemalbumin at 1.9Å. Biochem. Biophys. Res. Comm. 2002, 291, 813–819. [Google Scholar] [CrossRef]
- Chaves, O.A.; Amorim, A.P.O.; Castro, L.H.E.; Sant’Anna, C.M.R.; Oliveira, M.C.C.; Cesarin-Sobrinho, D.; Netto-Ferreira, J.C.; Ferreira, A.B.B. Fluorescence and docking studies of the interaction between human serum albumin and pheophytin. Molecules 2015, 20, 19526–19539. [Google Scholar] [CrossRef] [Green Version]
- Chaves, O.A.; Santos, M.R.L.; Oliveira, M.C.C.; Sant’Anna, C.M.R.; Ferreira, R.C.; Echevarria, A.; Netto-Ferreira, J.C. Synthesis, tyrosinase inhibition and transportation behavior of novel β-enamino thiosemicarbazide derivatives by human serum albumin. J. Mol. Liq. 2018, 254, 280–290. [Google Scholar] [CrossRef]
- Chaves, O.A.; Tavares, M.T.; Cunha, M.R.; Parise-Filho, R.; Sant’Anna, C.M.R.; Netto-Ferreira, J.C. Multi-spectroscopic and theoretical analysis on the interaction between human serum albumin and a capsaicin derivative—RPF101. Biomolecules 2018, 8, 78. [Google Scholar] [CrossRef] [Green Version]
- Britto, M.M.; Almeida, T.M.G.; Leitão, A.; Donnici, C.L.; Lopes, M.T.P.; Montanari, C.A. Synthesis of mesoionic 4-(para-substituted phenyl-5-2,4-dichlorophenyl)-1,3-4-thiadiazolium-2-aminides by direct cyclization via acylation of thiosemicarbazides. Synthetic Commun. 2006, 36, 3359–3369. [Google Scholar] [CrossRef]
- Sousa-Pereira, D.; Chaves, O.A.; Reis, C.M.; Oliveira, M.C.C.; Sant’Anna, C.M.R.; Netto-Ferreira, J.C.; Echevarria, A. Synthesis and biological evaluation of N-aryl-2-phenyl-hydrazinecarbothioamides: Experimental and theoretical analysis on tyrosinase inhibition and interaction with HSA. Bioorg. Chem. 2018, 81, 79–87. [Google Scholar] [CrossRef]
- Anderson, W.K.; Corey, P.F. Synthesis and antileukemic activity of 5-substituted 2,3-dihydro-6,7-bis(hydroxymethyl)-1H-pyrrolizine diesters. J. Med. Chem. 1977, 20, 812–818. [Google Scholar]
- Anderson, W.K.; Heider, A.R.; Raju, N.; Yucht, J.A. Synthesis and antileukemic activity of bis[[(carbamoyl)oxy]methyl]- substituted pyrrolo[2,1-a]isoquinolines, pyrrolo[1,2-a]quinolines, pyrrolo[2,1-a]isobenzazepines, and pyrrolo[1,2-a]benzazepines. J. Med. Chem. 1988, 31, 2097–2102. [Google Scholar] [CrossRef] [PubMed]
- Vilpo, J.A.; Vilpo, L.M.; Vuorinen, P.; Moilanen, E.; Metsä-Ketelä, T. Mode of cytostatic action of mesoionic oxatriazole nitric oxide donors in proliferating human hematopoietic cells. Anticancer Drug Des. 1997, 12, 75–89. [Google Scholar] [PubMed]
- Hillard, E.A.; Pigeon, P.; Vessières, A.; Amatore, C.; Jaouen, G. The influence of phenolic hydroxy substitution on the electron transfer and anti-cancer properties of compounds based on the 2-ferrocenyl-1-phenyl-but-1-ene motif. Dalton Trans. 2007, 43, 5073–5081. [Google Scholar]
- Gnanasekaran, K.K.; Pouland, T.; Bunce, R.A.; Darrell Berlin, K.; Abuskhuna, S.; Bhandari, D.; Mashayekhi, M.; Zhou, D.H.; Benbrook, D.M. Tetrahydroquinoline units in flexible heteroarotinoids (Flex-Hets) convey anti-cancer properties in A2780 ovarian cancer cells. Bioorg. Med. Chem. 2020, 28, 115244. [Google Scholar]
- Lynn, W.S.; Tweedale, A.; Cloyd, M.W. Human Immunodeficiency Virus (HIV-1) cytotoxicity: Perturbation of the cell membrane and depression of phospholipid synthesis. Virology 1988, 163, 43–51. [Google Scholar] [CrossRef]
- Olière, S.; Douville, R.; Sze, A.; Belgnaoui, S.M.; Hiscott, J. Modulation of innate immune responses during Human T-Cell Leukemia Virus (HTLV-1) pathogenesis. Cytokine Growth Factor Rev. 2011, 22, 197–210. [Google Scholar]
- Glaser, R.; Wu, H.; Lewis, M. Cytosine catalysis of nitrosative guanine deamination and interstrand cross-link formation. J. Am. Chem. Soc. 2005, 127, 7346–7358. [Google Scholar] [CrossRef]
- Gorrini, C.; Harris, I.S.; Mak, T.W. Modulation of oxidative stress as an anticancer strategy. Nat. Rev. Drug Discov 2013, 12, 931–947. [Google Scholar] [CrossRef]
- Lähteenmäki, T.; Sievi, E.; Vapaatalo, H. Inhibitory effects of mesoionic 3-aryl substituted oxatriazole-5-imine derivatives on vascular smooth muscle cell mitogenesis and proliferation in vitro. Br. J. Pharmacol. 1998, 125, 402–408. [Google Scholar]
- Naveenraj, S.; Anandan, S. Binding of serum albumins with bioactive substances – Nanoparticles to drugs. J. Photochem. Photobiol. C 2013, 14, 53–71. [Google Scholar]
- Koly, S.F.; Kundu, S.P.; Kabir, S.; Amran, M.S.; Sultan, M.Z. Analysis of aceclofenac and bovine serum albumin interaction using fluorescence quenching method for predictive, preventive, and personalized medicine. EPMA J. 2015, 6, 24–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montalti, M.; Credi, A.; Prodi, L.; Gandolfi, M.T. Handbook of Photochemistry, 3rd ed.; CRC Press, Taylor & Francis: Boca Raton, FL, USA, 2006. [Google Scholar]
- Millar, D.P. Time-resolved fluorescence spectroscopy. Curr. Opin. Struct. Biol. 1996, 6, 637–928. [Google Scholar] [PubMed]
- Mondal, M.; Lakshmi, P.T.; Krishna, R.; Sakthivel, N. Molecular interaction between human serum albumin (HSA) and phloroglucinol derivative that shows selective anti-proliferative potential. J. Lumin. 2017, 192, 990–998. [Google Scholar] [CrossRef]
- Lopes, N.D.; Chaves, O.A.; de Oliveira, M.C.C.; Sant’Anna, C.M.R.; Sousa-Pereira, D.; Netto-Ferreira, J.C.; Echevarria, A. Novel piperonal 1,3,4-thiadiazolium-2-phenylamines mesoionic derivatives: Synthesis, tyrosinase inhibition evaluation and HSA binding study. Int. J. Biol. Macromol. 2018, 112, 1062–1072. [Google Scholar] [CrossRef]
- Ross, P.D.; Subramanian, S. Thermodynamics of protein association reactions: Forces contributing to stability. Biochemistry 1981, 20, 3096–3102. [Google Scholar] [CrossRef]
- Chaves, O.A.; Jesus, C.S.H.; Henriques, E.S.; Brito, R.M.M.; Serpa, C. In-situ ultra-fast heat deposition does not perturb serum albumin structure. Photochem. Photobiol. Sci. 2016, 15, 1524–1535. [Google Scholar] [CrossRef]
- Chaves, O.A.; Mathew, B.; Cesarin-Sobrinho, D.; Lakshminarayanan, B.; Joy, M.; Mathew, G.E.; Suresh, J.; Netto-Ferreira, J.C. Spectroscopic, zeta potential and molecular docking analysis on the interaction between human serum albumin and halogenated thienyl chalcones. J. Mol. Liq. 2017, 242, 1018–1026. [Google Scholar] [CrossRef]
- Matei, I.; Hillebrand, M. Interaction of kaempferol with human serum albumin: A fluorescence and circular dichroism study. J. Pharm. Biomed. Anal. 2010, 51, 768–773. [Google Scholar] [CrossRef]
- Qian, Y.; Zhou, X.; Chen, J.; Zhang, Y. Binding of bezafibrate to human serum albumin: Insight into the non-covalent interaction of an emerging contaminant with biomacromolecules. Molecules 2012, 17, 6821–6831. [Google Scholar]
- Hosainzadeh, A.; Gharanfoli, M.; Saberi, M.R.; Chamani, J.K. Probing the interaction of human serum albumin with bilirubin in the presence of aspirin by multi-spectroscopic, molecular modeling and zeta potential techniques: Insight on binary and ternary systems. J. Biomol. Struct. Dyn. 2012, 29, 1013–1050. [Google Scholar] [CrossRef] [PubMed]
- Shen, G.-F.; Liu, T.-T.; Wang, Q.; Jiang, M.; Shi, J.-H. Spectroscopic and molecular docking studies of binding interaction of gefitinib, lapatinib and sunitinib with bovine serum albumin (BSA). J. Photochem. Photobiol. B 2015, 153, 380–390. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.-Y.; Zhang, H.-X.; Mezei, M.; Cui, M. Molecular docking: A powerful approach for structure-based drug discovery. Curr. Comput.-Aided Drug Des. 2011, 7, 146–157. [Google Scholar] [CrossRef] [PubMed]
- Sudlow, G.; Birkett, D.J.; Wade, D.N. The characterization of two specific drug binding sites on human serum albumin. Mol. Pharmacol. 1975, 11, 824–832. [Google Scholar]
- Sudlow, G.; Birkett, D.J.; Wade, D.N. Further characterization of specific drug binding sites on human serum albumin. Mol. Pharmacol. 1976, 12, 1052–1061. [Google Scholar]
- Yamasaki, K.; Chuang, V.T.G.; Maruyama, T.; Otagiri, M. Albumin-drug interaction and its clinical implication. Biochim. Biophys. Acta 2013, 1830, 5435–5443. [Google Scholar] [CrossRef]
- Chuang, V.T.; Otagiri, M. Flunitrazepam, a 7-nitro-1,4-benzodiazepine that is unable to bind to the indole-benzodiazepine site of human serum albumin. Biochim. Biophys. Acta 2001, 1546, 337–345. [Google Scholar] [CrossRef]
- Hehre, W.J. A guide to molecular mechanics and quantum chemical calculations. In Wavefunction; INC.: Irvine, CA, USA, 2003. [Google Scholar]
- Drew, H.R.; Wing, R.M.; Takano, T.; Broka, C.; Tanaka, S.; Itakura, K.; Dickerson, R.E. Structure of a B-DNA dodecamer: Conformation and dynamics. Proc. Natl. Acad. Sci. USA 1981, 78, 2179–2183. [Google Scholar] [CrossRef] [Green Version]
- GOLD-Protein Ligand Docking Software. Available online: http://www.ccdc.cam.ac.uk/solutions/csd-discovery/components/gold/ (accessed on 30 April 2020).
- DeLano, W.L. PyMOL User’s Guide; DeLano Scientific LLC: San Carlos, CA, USA, 2002. [Google Scholar]
Sample Availability: Samples are available from the authors. |











| Cell Lines | IC50 Values for the Mesoionic Compounds 5a–d (μM) | |||
|---|---|---|---|---|
| 5a | 5b | 5c | 5d | |
| MT2 | 1.51 | 5.09 | 6.15 | 7.70 |
| C91/PL | 2.82 | 4.50 | 4.50 | 5.25 |
| Jurkat | 2.15 | 1.74 | 5.60 | 2.27 |
| Sample | T (K) | KSV (× 105) (M−1) | kq (× 1013) (M−1s−1) | Ka (× 104) (M−1) | ΔH° (kJ.mol−1) | ΔS° (× 10−2) (kJ.mol−1K−1) | ΔG° (kJ.mol−1) |
|---|---|---|---|---|---|---|---|
| 296 | 1.19 ± 0.03 | 2.15 | 7.68 ± 0.26 | −27.7 | |||
| HSA:5a | 303 | 1.20 ± 0.03 | 2.16 | 7.51 ± 0.26 | −2.64 ± 0.15 | 8.46 ± 0.05 | −28.3 |
| 310 | 1.21 ± 0.03 | 2.18 | 7.32 ± 0.26 | −28.9 | |||
| 296 | 1.10 ± 0.02 | 1.99 | 8.57 ± 0.26 | −28.0 | |||
| HSA:5b | 303 | 1.14 ± 0.03 | 2.06 | 8.44 ± 0.26 | −1.88 ± 0.17 | 8.81 ± 0.06 | −28.6 |
| 310 | 1.15 ± 0.03 | 2.07 | 8.27 ± 0.26 | −29.2 | |||
| 296 | 1.30 ± 0.03 | 2.35 | 8.53 ± 0.26 | −28.1 | |||
| HSA:5c | 303 | 1.36 ± 0.04 | 2.45 | 9.06 ± 0.26 | 6.27 ± 0.10 | 11.6 ± 0.1 | −28.9 |
| 310 | 1.40 ± 0.03 | 2.53 | 9.57 ± 0.26 | −29.7 | |||
| 296 | 1.45 ± 0.02 | 2.61 | 8.71 ± 0.26 | −27.9 | |||
| HSA:5d | 303 | 1.54 ± 0.03 | 2.79 | 9.02 ± 0.26 | 3.43 ± 0.16 | 10.6 ± 0.1 | −28.7 |
| 310 | 1.60 ± 0.03 | 2.89 | 9.28 ± 0.26 | −29.4 |
| Sample | τ1 (ns) | Relative % | τ2 (ns) | Relative % | χ2 |
|---|---|---|---|---|---|
| HSA | 1.73 ± 0.16 | 20.7 | 5.54 ± 0.12 | 79.3 | 1.095 |
| HSA:5a | 1.61 ± 0.13 | 26.4 | 5.38 ± 0.13 | 73.6 | 1.140 |
| HSA:5b | 1.66 ± 0.12 | 23.9 | 5.25 ± 0.11 | 76.1 | 1.106 |
| HSA:5c | 1.68 ± 0.11 | 33.0 | 5.66 ± 0.16 | 67.0 | 1.155 |
| HSA:5d | 1.65 ± 0.10 | 29.8 | 5.37 ± 0.13 | 70.2 | 1.038 |
| CD Results | ZP Results | ||||
|---|---|---|---|---|---|
| Sample | 208 nm | 222 nm | ζ (mV) | Conductance (µs) | Current (mA) |
| HSA | 86.7% | 80.1% | −9.15 ± 1.79 | 29,645 | 150 |
| HSA:5a | 84.0% | 78.5% | −8.62 ± 1.40 | 29,615 | 150 |
| HSA:5b | 82.4% | 77.3% | −8.55 ± 1.46 | 29,642 | 150 |
| HSA:5c | 83.8% | 78.8% | −8.77 ± 1.52 | 29,622 | 150 |
| HSA:5d | 82.0% | 77.7% | −8.69 ± 1.50 | 29,638 | 150 |
| DNA | HSA | ||||
|---|---|---|---|---|---|
| Compound | Minor Groove | Major Groove | Site I | Site II | Site III |
| 5a | 83.9 | 11.2 | 78.3 | 40.2 | 39.5 |
| 5b | 84.8 | 10.6 | 80.1 | 40.0 | 38.1 |
| 5c | 83.4 | 13.8 | 86.2 | 45.6 | 41.1 |
| 5d | 87.4 | 13.6 | 85.0 | 42.9 | 40.8 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sousa-Pereira, D.; Silva de Oliveira, T.; Paiva, R.O.; Chaves, O.A.; Netto-Ferreira, J.C.; Echevarria-Lima, J.; Echevarria, A. Synthetic (E)-3-Phenyl-5-(phenylamino)-2-styryl-1,3,4-thiadiazol-3-ium Chloride Derivatives as Promising Chemotherapy Agents on Cell Lines Infected with HTLV-1. Molecules 2020, 25, 2537. https://doi.org/10.3390/molecules25112537
Sousa-Pereira D, Silva de Oliveira T, Paiva RO, Chaves OA, Netto-Ferreira JC, Echevarria-Lima J, Echevarria A. Synthetic (E)-3-Phenyl-5-(phenylamino)-2-styryl-1,3,4-thiadiazol-3-ium Chloride Derivatives as Promising Chemotherapy Agents on Cell Lines Infected with HTLV-1. Molecules. 2020; 25(11):2537. https://doi.org/10.3390/molecules25112537
Chicago/Turabian StyleSousa-Pereira, Danilo, Thais Silva de Oliveira, Rojane O. Paiva, Otávio Augusto Chaves, José C. Netto-Ferreira, Juliana Echevarria-Lima, and Aurea Echevarria. 2020. "Synthetic (E)-3-Phenyl-5-(phenylamino)-2-styryl-1,3,4-thiadiazol-3-ium Chloride Derivatives as Promising Chemotherapy Agents on Cell Lines Infected with HTLV-1" Molecules 25, no. 11: 2537. https://doi.org/10.3390/molecules25112537