Comparative Transcriptomic Analyses Provide Insights into the Enzymatic Browning Mechanism of Fresh-Cut Sand Pear Fruit
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Browning Measurement in Sand Pear Flesh Slices
2.3. RNA Extraction
2.4. SMRT, Illumina RNA Sequencing, and Data Analysis
2.5. Quantitative Real-Time PCR (qRT-PCR) Analysis
2.6. Measurement of Total Phenolic Content and Monomeric Compounds
2.7. Assessment of Enzyme Activity
3. Results
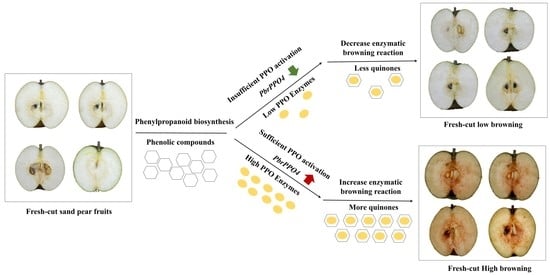
3.1. Flesh Browning Intensity
3.2. SMRT- and Illumina RNA-seq
3.3. Identification of AS and lncRNAs
3.4. Analysis of DEGs and qRT-PCR Confirmation
3.5. Annotation and Enrichment Analysis of DEGs
3.6. Phenylpropanoid and Flavonoid Biosynthesis Related Gene Dynamics and Polyphenol Quantification
3.7. PPO and POD Gene Expression and Enzyme Activities
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Friedman, M. Food browning and its prevention: An overview. J. Agric. Food Chem. 1996, 44, 631–653. [Google Scholar] [CrossRef]
- Gomes, M.H.; Vieira, T.; Fundo, J.F.; Almeida, D.P. Polyphenoloxidase activity and browning in fresh-cut ‘Rocha’ pear as affected by pH, phenolic substrates, and antibrowning additives. Postharvest Biol. Technol. 2014, 91, 32–38. [Google Scholar] [CrossRef] [Green Version]
- Cheng, Y.; Liu, L.; Feng, Y.; Dong, Y.; Guan, J. Effects of 1-MCP on fruit quality and core browning in ‘Yali’ pear during cold storage. Sci. Hortic. 2019, 243, 350–356. [Google Scholar] [CrossRef]
- Liao, T.; Liu, J.; Sun, Y.; Zou, L.; Zhou, L.; Liu, C.; Terefe, N.S.; Liu, W. Differential inhibitory effects of organic acids on pear polyphenol oxidase in model systems and pear puree. LWT Food Sci. Technol. 2020, 118, 108704. [Google Scholar] [CrossRef]
- Terefe, N.S.; Buckow, R.; Versteeg, C. Quality-related enzymes in fruit and vegetable products: Effects of novel food processing technologies, part 1: High-pressure processing. Crit. Rev. Food Sci. Nutr. 2014, 54, 24–63. [Google Scholar] [CrossRef] [PubMed]
- Mayer, A.M. Polyphenol oxidases in plants and fungi: Going places? A review. Phytochemistry 2006, 67, 2318–2331. [Google Scholar] [CrossRef] [PubMed]
- Tran, L.T.; Taylor, J.S.; Constabel, C.P. The polyphenol oxidase gene family in land plants: Lineage-specific duplication and expansion. BMC Genom. 2012, 13, 395. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Sun, X. Recent advances in polyphenol oxidase-mediated plant stress responses. Phytochemistry 2021, 181, 112588. [Google Scholar] [CrossRef]
- Chi, M.; Bhagwat, B.; Lane, W.D.; Tang, G.; Su, Y.; Sun, R.; Oomah, B.D.; Wiersma, P.A.; Xiang, Y. Reduced polyphenol oxidase gene expression and enzymatic browning in potato (Solanum tuberosum L.) with artificial microRNAs. BMC Plant. Biol. 2014, 14, 62. [Google Scholar] [CrossRef] [Green Version]
- González, M.N.; Massa, G.A.; Andersson, M.; Turesson, H.; Olsson, N.; Fält, A.-S.; Storani, L.; Décima Oneto, C.A.; Hofvander, P.; Feingold, S.E. Reduced enzymatic browning in potato tubers by specific editing of a polyphenol oxidase gene via ribonucleoprotein complexes delivery of the CRISPR/Cas9 system. Front. Plant. Sci. 2020, 10, 1649. [Google Scholar] [CrossRef] [PubMed]
- Di Guardo, M.; Tadiello, A.; Farneti, B.; Lorenz, G.; Masuero, D.; Vrhovsek, U.; Costa, G.; Velasco, R.; Costa, F. A multidisciplinary approach providing new insight into fruit flesh browning physiology in apple (Malus x domestica Borkh.). PLoS ONE 2013, 8, e78004. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Meng, S.; Xiang, Z.; Yang, G.; He, N. An R1R2R3 MYB transcription factor, MnMYB3R1, regulates the polyphenol oxidase gene in mulberry (Morus notabilis). Int. J. Mol. Sci. 2019, 20, 2602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, H.; Chen, C.; Zeng, J.; Yun, Z.; Liu, Y.; Qu, H.; Jiang, Y.; Duan, X.; Xia, R. Micro RNA 528, a hub regulator modulating ROS homeostasis via targeting of a diverse set of genes encoding copper-containing proteins in monocots. New Phytol. 2020, 225, 385–399. [Google Scholar] [CrossRef]
- Taranto, F.; Pasqualone, A.; Mangini, G.; Tripodi, P.; Miazzi, M.M.; Pavan, S.; Montemurro, C. Polyphenol oxidases in crops: Biochemical, physiological and genetic aspects. Int. J. Mol. Sci. 2017, 18, 377. [Google Scholar] [CrossRef]
- Tanrıöven, D.; Ekşi, A. Phenolic compounds in pear juice from different cultivars. Food Chem. 2005, 93, 89–93. [Google Scholar] [CrossRef]
- Li, X.; Wang, T.; Zhou, B.; Gao, W.; Cao, J.; Huang, L. Chemical composition and antioxidant and anti-inflammatory potential of peels and flesh from 10 different pear varieties (Pyrus spp.). Food Chem. 2014, 152, 531–538. [Google Scholar] [CrossRef] [PubMed]
- Kolniak-Ostek, J. Identification and quantification of polyphenolic compounds in ten pear cultivars by UPLC-PDA-Q/TOF-MS. J. Food Compos. Anal. 2016, 49, 65–77. [Google Scholar] [CrossRef]
- Li, X.; Zhang, J.-Y.; Gao, W.-Y.; Wang, Y.; Wang, H.-Y.; Cao, J.-G.; Huang, L.-Q. Chemical composition and anti-inflammatory and antioxidant activities of eight pear cultivars. J. Agric. Food Chem. 2012, 60, 8738–8744. [Google Scholar] [CrossRef]
- Wang, X.; Liu, C.; Wang, Q.; Liu, C.; Sun, H.; Zhang, M.; Lyu, D.; Du, G. A comparative study on antioxidant, anti-inflammatory, antimicrobial activities and chemical composition of Pyrus ussuriensis cultivars from northeastern China. Hortic. Environ. Biotechnol. 2021, 62, 477–491. [Google Scholar] [CrossRef]
- Wang, Z.; Barrow, C.J.; Dunshea, F.R.; Suleria, H.A. A comparative investigation on phenolic composition, characterization and antioxidant potentials of five different Australian grown pear varieties. Antioxidants 2021, 10, 151. [Google Scholar] [CrossRef]
- Zheng, H.; Liu, W.; Liu, S.; Liu, C.; Zheng, L. Effects of melatonin treatment on the enzymatic browning and nutritional quality of fresh-cut pear fruit. Food Chem. 2019, 299, 125116. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Liu, L.; Zhao, G.; Shen, C.; Yan, H.; Guan, J.; Yang, K. The effects of modified atmosphere packaging on core browning and the expression patterns of PPO and PAL genes in ‘Yali’ pears during cold storage. LWT Food Sci. Technol. 2015, 60, 1243–1248. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, J.-W.; Zhou, B.; Liu, Y.-F.; Xia, X.-L.; Xiao, Z.-G.; Fei, L.; Ji, S.-J. Calcium inhibited peel browning by regulating enzymes in membrane metabolism of ‘Nanguo’ pears during post-ripeness after refrigerated storage. Sci. Hortic. 2019, 244, 15–21. [Google Scholar] [CrossRef]
- Dixon, R.A.; Achnine, L.; Kota, P.; Liu, C.J.; Reddy, M.S.; Wang, L. The phenylpropanoid pathway and plant defence-a genomics perspective. Mol. Plant. Pathol. 2002, 3, 371–390. [Google Scholar] [CrossRef]
- MacLean, D.; Murr, D.; DeEll, J.; Mackay, A.; Kupferman, E. Inhibition of PAL, CHS, and ERS1 in ‘Red d’Anjou’ pear (Pyrus communis L.) by 1-MCP. Postharvest Biol. Technol. 2007, 45, 46–55. [Google Scholar] [CrossRef]
- Wang, J.; Dong, S.; Jiang, Y.; He, H.; Liu, T.; Lv, M.; Ji, S. Influence of long-term cold storage on phenylpropanoid and soluble sugar metabolisms accompanied with peel browning of ‘Nanguo’ pears during subsequent shelf life. Sci. Hortic. 2020, 260, 108888. [Google Scholar] [CrossRef]
- Vogt, T. Phenylpropanoid biosynthesis. Mol. Plant. 2010, 3, 2–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saeeduddin, M.; Abid, M.; Jabbar, S.; Wu, T.; Hashim, M.M.; Awad, F.N.; Hu, B.; Lei, S.; Zeng, X. Quality assessment of pear juice under ultrasound and commercial pasteurization processing conditions. LWT Food Sci. Technol. 2015, 64, 452–458. [Google Scholar] [CrossRef]
- Sharma, S.; Rao, T.R. Xanthan gum based edible coating enriched with cinnamic acid prevents browning and extends the shelf-life of fresh-cut pears. LWT Food Sci. Technol. 2015, 62, 791–800. [Google Scholar] [CrossRef]
- Zhou, X.; Xiao, Y.; Meng, X.; Liu, B. Full inhibition of Whangkeumbae pear polyphenol oxidase enzymatic browning reaction by L-cysteine. Food Chem. 2018, 266, 1–8. [Google Scholar] [CrossRef]
- Steinwand, M.A.; Ronald, P.C. Crop biotechnology and the future of food. Nat. Food 2020, 1, 273–283. [Google Scholar] [CrossRef]
- Maioli, A.; Gianoglio, S.; Moglia, A.; Acquadro, A.; Valentino, D.; Milani, A.M.; Prohens, J.; Orzaez, D.; Granell, A.; Lanteri, S. Simultaneous CRISPR/Cas9 editing of three PPO genes reduces fruit flesh browning in Solanum melongena L. Front. Plant. Sci. 2020, 11, 1883. [Google Scholar] [CrossRef]
- Wang, T.; Zhang, C.; Zhang, H.; Zhu, H. CRISPR/Cas9-mediated gene editing revolutionizes the improvement of horticulture food crops. J. Agric. Food Chem. 2021. [Google Scholar] [CrossRef]
- Liu, Z.; Cheng, K.; Qin, Z.; Wu, T.; Li, X.; Tu, J.; Yang, F.; Zhu, H.; Yang, L. Selection and validation of suitable reference genes for qRT-PCR analysis in pear leaf tissues under distinct training systems. PLoS ONE 2018, 13, e0202472. [Google Scholar] [CrossRef] [PubMed]
- Syed, N.H.; Kalyna, M.; Marquez, Y.; Barta, A.; Brown, J.W. Alternative splicing in plants-coming of age. Trends Plant. Sci. 2012, 17, 616–623. [Google Scholar] [CrossRef] [Green Version]
- Correa, J.P.d.O.; Silva, E.M.; Nogueira, F.T. Molecular control by non-coding RNAs during fruit development: From gynoecium patterning to fruit ripening. Front. Plant. Sci. 2018, 9, 1760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waseem, M.; Liu, Y.; Xia, R. Long non-coding RNAs, the dark matter: An emerging regulatory component in plants. Int. J. Mol. Sci. 2021, 22, 86. [Google Scholar] [CrossRef]
- Wang, B.; Tseng, E.; Regulski, M.; Clark, T.A.; Hon, T.; Jiao, Y.; Lu, Z.; Olson, A.; Stein, J.C.; Ware, D. Unveiling the complexity of the maize transcriptome by single-molecule long-read sequencing. Nat. Commun. 2016, 7, 11708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, B.; Lu, T.; Xu, Y.; Wang, R.; Chen, G. Identification of genes associated with ricinoleic acid accumulation in Hiptage benghalensis via transcriptome analysis. Biotechnol. Biofuels 2019, 12, 16. [Google Scholar] [CrossRef] [Green Version]
- Rao, S.; Yu, T.; Cong, X.; Xu, F.; Lai, X.; Zhang, W.; Liao, Y.; Cheng, S. Integration analysis of PacBio SMRT and Illumina RNA-seq reveals candidate genes and pathway involved in selenium metabolism in hyperaccumulator Cardamine violifolia. BMC Plant. Biol. 2020, 20, 492. [Google Scholar] [CrossRef]
- Yang, Q.; Wang, J.; Wang, Z.; Li, X.; Kan, J.; Lin, J. SMRT sequencing of full-length transcriptome of birch-leaf pear (Pyrus betulifolia Bunge) under drought stress. J. Genet. 2021, 100, 29. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lin, J.; Kan, J.; Wang, H.; Li, X.; Yang, Q.; Li, H.; Chang, Y. Genome-wide identification and functional prediction of novel drought-responsive lncRNAs in Pyrus betulifolia. Genes 2018, 9, 311. [Google Scholar] [CrossRef] [Green Version]
- Ye, J.; Cheng, S.; Zhou, X.; Chen, Z.; Kim, S.U.; Tan, J.; Zheng, J.; Xu, F.; Zhang, W.; Liao, Y. A global survey of full-length transcriptome of Ginkgo biloba reveals transcript variants involved in flavonoid biosynthesis. Ind. Crop. Prod. 2019, 139, 111547. [Google Scholar] [CrossRef]
- Mishra, B.B.; Gautam, S.; Sharma, A. Free phenolics and polyphenol oxidase (PPO): The factors affecting post-cut browning in eggplant (Solanum melongena). Food Chem. 2013, 139, 105–114. [Google Scholar] [CrossRef]
- Hamauzu, Y.; Yasui, H.; Inno, T.; Kume, C.; Omanyuda, M. Phenolic profile, antioxidant property, and anti-influenza viral activity of Chinese quince (Pseudocydonia sinensis Schneid.), quince (Cydonia oblonga Mill.), and apple (Malus domestica Mill.) fruits. J. Agric. Food Chem. 2005, 53, 928–934. [Google Scholar] [CrossRef]
- Amiot, M.J.; Tacchini, M.; Aubert, S.Y.; Oleszek, W. Influence of cultivar, maturity stage, and storage conditions on phenolic composition and enzymic browning of pear fruits. J. Agric. Food Chem. 1995, 43, 1132–1137. [Google Scholar] [CrossRef]
- Yang, H.; Tian, T.; Gu, H.; Li, X.; Cai, G.; Sun, J.; Wu, D.; Lu, J. Analysis of factors related to browning of Dangshan pear (Pyrus spp.) wine. Food Chem. 2020, 308, 125665. [Google Scholar] [CrossRef]
- Fan, M.; Li, W.; Hu, X.; Sun, Y.n.; Yu, G.; Zhang, X. Effect of micro-vacuum storage on active oxygen metabolism, internal browning and related enzyme activities in Laiyang pear (Pyrus bretschneideri Reld). LWT Food Sci.Technol. 2016, 72, 467–474. [Google Scholar] [CrossRef]
- Tang, T.; Xie, X.; Ren, X.; Wang, W.; Tang, X.; Zhang, J.; Wang, Z. A difference of enzymatic browning unrelated to PPO from physiology, targeted metabolomics and gene expression analysis in Fuji apples. Postharvest Biol. Technol. 2020, 170, 111323. [Google Scholar] [CrossRef]
- Zhang, S.; Tian, L.; Zhang, Y.; Zhao, H.; Zhao, J.; Guo, J.; Zhu, G.-P. De novo transcriptome assembly of the fresh-cut white husk of Juglans cathayensis Dode: Insights for enzymatic browning mechanism of fresh-cut husk of walnut. Sci. Hortic. 2019, 257, 108654. [Google Scholar] [CrossRef]
- Huang, S.-J.; Lin, S.-Y.; Wang, T.-T.; Hsu, F.-C. Combining acetic acid and ethanol as an anti-browning treatment for lettuce butt discoloration through repression of the activity and expression of phenylalanine ammonia lyase. Postharvest Biol. Technol. 2020, 164, 111151. [Google Scholar] [CrossRef]
- Worarad, K.; Suzuki, T.; Norii, H.; Mochizuki, Y.; Ishii, T.; Shinohara, K.; Miyamoto, T.; Kuboyama, T.; Inoue, E. Transcriptome profiling for pericarp browning during long-term storage of intact lotus root (Nelumbo nucifera). Plant. Growth Regul. 2021, 95, 207–221. [Google Scholar] [CrossRef]
- Zhou, H.; Tian, M.; Huang, W.; Luo, S.; Hu, H.; Zhang, Y.; Zhang, L.; Li, P. Physiological and transcriptomic analysis of ‘Whangkeumbae’ pear core browning during low-temperature storage. Gene Expr. Patterns 2020, 36, 119113. [Google Scholar] [CrossRef] [PubMed]






| Compound | Samples | |||
|---|---|---|---|---|
| LB-0h | LB-1h | HB-0h | HB-1h | |
| Total polyphenolic | 117 ± 11 a | 118 ± 10 a | 683 ± 34 b * | 503 ± 38 c |
| Chlorogenic acid | 0.17 ± 0.1 a | 0.12 ± 0.1 a | 78.1 ± 9.8 b * | 42.9 ± 9.3 c |
| Catechin | ND | ND | ND | ND |
| Epicatechin | ND | ND | ND | ND |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, J.; Du, W.; Chen, Q.-L.; Zhang, J.-G.; Yang, X.-P.; Hussain, S.B.; Hu, H.-J. Comparative Transcriptomic Analyses Provide Insights into the Enzymatic Browning Mechanism of Fresh-Cut Sand Pear Fruit. Horticulturae 2021, 7, 502. https://doi.org/10.3390/horticulturae7110502
Fan J, Du W, Chen Q-L, Zhang J-G, Yang X-P, Hussain SB, Hu H-J. Comparative Transcriptomic Analyses Provide Insights into the Enzymatic Browning Mechanism of Fresh-Cut Sand Pear Fruit. Horticulturae. 2021; 7(11):502. https://doi.org/10.3390/horticulturae7110502
Chicago/Turabian StyleFan, Jing, Wei Du, Qi-Liang Chen, Jing-Guo Zhang, Xiao-Ping Yang, Syed Bilal Hussain, and Hong-Ju Hu. 2021. "Comparative Transcriptomic Analyses Provide Insights into the Enzymatic Browning Mechanism of Fresh-Cut Sand Pear Fruit" Horticulturae 7, no. 11: 502. https://doi.org/10.3390/horticulturae7110502