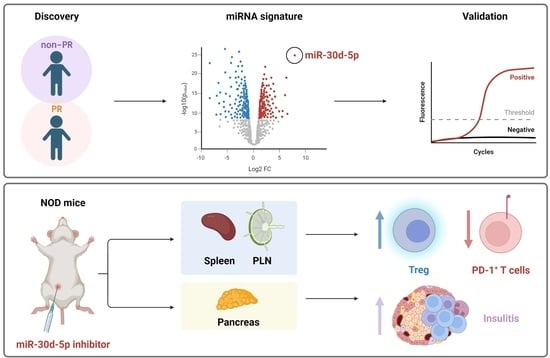
Immunoregulatory Biomarkers of the Remission Phase in Type 1 Diabetes: miR-30d-5p Modulates PD-1 Expression and Regulatory T Cell Expansion
Abstract
:1. Introduction
2. Results
2.1. Distinct miRNA Signature during the Partial Remission Phase of Pediatric Patients with Type 1 Diabetes
2.2. Validation of Differentially Expressed miRNAs during the Partial Remission Phase
2.3. Potential Enriched Biological Functions and Pathways under the Regulation of Differentially Expressed miRNAs during the Partial Remission Phase
2.4. In Vivo Inhibition of miR-30d-5p Potentiates Treg Expansion by Increasing CD200 Levels
2.5. In Vivo Inhibition of miR-30d-5p Reduces PD-1 Expression on Splenocytes
2.6. In Vivo Inhibition of miR-30d-5p Displays Changes in Additional T Cell Differentiation Subsets
2.7. In Vivo Inhibition of miR-30d-5p Tends to Increase Leukocyte Islet Infiltration
3. Discussion
4. Materials and Methods
4.1. Human Sample Collection and T1D Remission Follow-Up
4.2. Clinical and Laboratory Testing
4.3. Total RNA Isolation
4.4. RNA Library Preparation, Sequencing, and Data Analysis
4.5. Gene Targets for miRNAs
4.6. Gene Ontology and Pathway Analysis
4.7. Quantitative RT-qPCR
4.8. Mice
4.9. In Vivo miR-30d-5p Inhibitor Administration
4.10. Insulitis Score
4.11. Flow Cytometry
4.11.1. Leukocyte Isolation from Spleen and PLN
4.11.2. Viability and Cell Counting
4.11.3. Immunophenotype
4.12. Statistical Analysis
4.13. Ethics
4.13.1. Human Samples
4.13.2. Mice
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Patterson, C.C.; Harjutsalo, V.; Rosenbauer, J.; Neu, A.; Cinek, O.; Skrivarhaug, T.; Rami-Merhar, B.; Soltesz, G.; Svensson, J.; Parslow, R.C.; et al. Trends and cyclical variation in the incidence of childhood type 1 diabetes in 26 European centres in the 25 year period 1989–2013: A multicentre prospective registration study. Diabetologia 2018, 62, 408–417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Megen, K.M.; Spindler, M.P.; Keij, F.M.; Bosch, I.; Sprangers, F.; van Royen-Kerkhof, A.; Nikolic, T.; Roep, B.O. Relapsing/remitting type 1 diabetes. Diabetologia 2017, 60, 2252–2255. [Google Scholar] [CrossRef] [Green Version]
- Von Herrath, M.; Sanda, S.; Herold, K.C. Type 1 diabetes as a relapsing–remitting disease? Nat. Rev. Immunol. 2007, 7, 988–994. [Google Scholar] [CrossRef] [PubMed]
- Fonolleda, M.; Murillo, M.; Vázquez, F.; Bel, J.; Vives-Pi, M. Remission Phase in Paediatric Type 1 Diabetes: New Understanding and Emerging Biomarkers. Horm. Res. Paediatr. 2017, 88, 307–315. [Google Scholar] [CrossRef] [Green Version]
- Tang, R.; Zhong, T.; Wu, C.; Zhou, Z.; Li, X. The Remission Phase in Type 1 Diabetes: Role of Hyperglycemia Rectification in Immune Modulation. Front. Endocrinol. 2019, 10, 824. [Google Scholar] [CrossRef] [Green Version]
- Karges, B.; Durinovic-Belló, I.; Heinze, E.; Debatin, K.-M.; Boehm, B.; Karges, W. Immunological mechanisms associated with long-term remission of human type 1 diabetes. Diabetes/Metabolism Res. Rev. 2005, 22, 184–189. [Google Scholar] [CrossRef]
- Fitas, A.L.; Martins, C.; Borrego, L.M.; Lopes, L.; Jörns, A.; Lenzen, S.; Limbert, C. Immune cell and cytokine patterns in children with type 1 diabetes mellitus undergoing a remission phase: A longitudinal study. Pediatr. Diabetes 2018, 19, 963–971. [Google Scholar] [CrossRef]
- Gomez-Muñoz, L.; Perna-Barrull, D.; Villalba, A.; Rodriguez-Fernandez, S.; Ampudia, R.-M.; Teniente-Serra, A.; Vazquez, F.; Murillo, M.; Perez, J.; Corripio, R.; et al. NK Cell Subsets Changes in Partial Remission and Early Stages of Pediatric Type 1 Diabetes. Front. Immunol. 2021, 11, 3551. [Google Scholar] [CrossRef] [PubMed]
- Narsale, A.; Lam, B.; Moya, R.; Lu, T.; Mandelli, A.; Gotuzzo, I.; Pessina, B.; Giamporcaro, G.; Geoffrey, R.; Buchanan, K.; et al. CD4+CD25+CD127hi cell frequency predicts disease progression in type 1 diabetes. J. Clin. Investig. 2021, 6, e136114. [Google Scholar] [CrossRef]
- Schloot, N.C.; Hanifi-Moghaddam, P.; Aabenhus-Andersen, N.; Alizadeh, B.Z.; Saha, M.T.; Knip, M.; Devendra, D.; Wilkin, T.; Bonifacio, E.; Roep, B.O.; et al. Association of immune mediators at diagnosis of Type 1 diabetes with later clinical remission. Diabet. Med. 2007, 24, 512–520. [Google Scholar] [CrossRef]
- Long, S.A.; Thorpe, J.; DeBerg, H.A.; Gersuk, V.; Eddy, J.A.; Harris, K.M.; Ehlers, M.; Herold, K.C.; Nepom, G.T.; Linsley, P.S. Partial exhaustion of CD8 T cells and clinical response to teplizumab in new-onset type 1 diabetes. Sci. Immunol. 2016, 1, eaai7793. [Google Scholar] [CrossRef] [Green Version]
- Arif, S.; Leete, P.; Nguyen, V.; Marks, K.; Nor, N.M.; Estorninho, M.; Kronenberg-Versteeg, D.; Bingley, P.J.; Todd, J.A.; Guy, C.; et al. Blood and Islet Phenotypes Indicate Immunological Heterogeneity in Type 1 Diabetes. Diabetes 2014, 63, 3835–3845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leete, P.; Willcox, A.; Krogvold, L.; Dahl-Jørgensen, K.; Foulis, A.K.; Richardson, S.J.; Morgan, N.G. Differential Insulitic Profiles Determine the Extent of β-Cell Destruction and the Age at Onset of Type 1 Diabetes. Diabetes 2016, 65, 1362–1369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomez-Muñoz, L.; Perna-Barrull, D.; Caroz-Armayones, J.M.; Murillo, M.; Rodriguez-Fernandez, S.; Valls, A.; Vazquez, F.; Perez, J.; Corripio, R.; Castaño, L.; et al. Candidate Biomarkers for the Prediction and Monitoring of Partial Remission in Pediatric Type 1 Diabetes. Front. Immunol. 2022, 13, 555. [Google Scholar] [CrossRef]
- Li, X.; Zhong, T.; Tang, R.; Wu, C.; Xie, Y.; Liu, F.; Zhou, Z. PD-1 and PD-L1 Expression in Peripheral CD4/CD8+ T Cells Is Restored in the Partial Remission Phase in Type 1 Diabetes. J. Clin. Endocrinol. Metab. 2020, 105, 1947–1956. [Google Scholar] [CrossRef] [PubMed]
- Villalba, A.; Fonolleda, M.; Murillo, M.; Rodriguez-Fernandez, S.; Ampudia, R.-M.; Perna-Barrull, D.; Raina, M.B.; Quirant-Sanchez, B.; Planas, R.; Teniente-Serra, A.; et al. Partial remission and early stages of pediatric type 1 diabetes display immunoregulatory changes. A pilot study. Transl. Res. 2019, 210, 8–25. [Google Scholar] [CrossRef] [Green Version]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Paul, P.; Chakraborty, A.; Sarkar, D.; Langthasa, M.; Rahman, M.; Bari, M.; Singha, R.S.; Malakar, A.K.; Chakraborty, S. Interplay between miRNAs and human diseases. J. Cell. Physiol. 2017, 233, 2007–2018. [Google Scholar] [CrossRef] [PubMed]
- Jin, F.; Hu, H.; Xu, M.; Zhan, S.; Wang, Y.; Zhang, H.; Chen, X. Serum microRNA Profiles Serve as Novel Biomarkers for Autoimmune Diseases. Front. Immunol. 2018, 9, 2381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simpson, L.J.; Ansel, K.M. MicroRNA regulation of lymphocyte tolerance and autoimmunity. J. Clin. Investig. 2015, 125, 2242–2249. [Google Scholar] [CrossRef] [Green Version]
- Xu, G.; Thielen, L.A.; Chen, J.; Grayson, T.B.; Grimes, T.; Bridges, S.L.; Tse, H.M.; Smith, B.; Patel, R.; Li, P.; et al. Serum miR-204 is an early biomarker of type 1 diabetes-associated pancreatic beta-cell loss. Am. J. Physiol. Metab. 2019, 317, E723–E730. [Google Scholar] [CrossRef] [PubMed]
- Scherm, M.G.; Daniel, C. miRNA-Mediated Immune Regulation in Islet Autoimmunity and Type 1 Diabetes. Front. Endocrinol. 2020, 11, 606322. [Google Scholar] [CrossRef]
- Grieco, F.A.; Sebastiani, G.; Juan-Mateu, J.; Villate, O.; Marroqui, L.; Ladrière, L.; Tugay, K.; Regazzi, R.; Bugliani, M.; Marchetti, P.; et al. MicroRNAs miR-23a-3p, miR-23b-3p, and miR-149-5p Regulate the Expression of Proapoptotic BH3-Only Proteins DP5 and PUMA in Human Pancreatic β-Cells. Diabetes 2016, 66, 100–112. [Google Scholar] [CrossRef] [Green Version]
- Guay, C.; Kruit, J.K.; Rome, S.; Menoud, V.; Mulder, N.L.; Jurdzinski, A.; Mancarella, F.; Sebastiani, G.; Donda, A.; Gonzalez, B.J.; et al. Lymphocyte-Derived Exosomal MicroRNAs Promote Pancreatic β Cell Death and May Contribute to Type 1 Diabetes Development. Cell Metab. 2019, 29, 348–361.e6. [Google Scholar] [CrossRef] [Green Version]
- Scherm, M.G.; Serr, I.; Zahm, A.M.; Schug, J.; Bellusci, S.; Manfredini, R.; Salb, V.K.; Gerlach, K.; Weigmann, B.; Ziegler, A.-G.; et al. miRNA142-3p targets Tet2 and impairs Treg differentiation and stability in models of type 1 diabetes. Nat. Commun. 2019, 10, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Erener, S.; Marwaha, A.; Tan, R.; Panagiotopoulos, C.; Kieffer, T.J. Profiling of circulating microRNAs in children with recent onset of type 1 diabetes. J. Clin. Investig. 2017, 2, e89656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samandari, N.; Mirza, A.H.; Kaur, S.; Hougaard, P.; Nielsen, L.B.; Fredheim, S.; Mortensen, H.B.; Pociot, F. Influence of Disease Duration on Circulating Levels of miRNAs in Children and Adolescents with New Onset Type 1 Diabetes. Non-Coding RNA 2018, 4, 35. [Google Scholar] [CrossRef] [Green Version]
- Assmann, T.S.; Recamonde-Mendoza, M.; Puñales, M.; Tschiedel, B.; Canani, L.H.; Crispim, D. MicroRNA expression profile in plasma from type 1 diabetic patients: Case-control study and bioinformatic analysis. Diabetes Res. Clin. Pract. 2018, 141, 35–46. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, L.B.; Wang, C.; Sørensen, K.; Bang-Berthelsen, C.H.; Hansen, L.; Andersen, M.-L.M.; Hougaard, P.; Juul, A.; Zhang, C.-Y.; Pociot, F.; et al. Circulating Levels of MicroRNA from Children with Newly Diagnosed Type 1 Diabetes and Healthy Controls: Evidence That miR-25 Associates to Residual Beta-Cell Function and Glycaemic Control during Disease Progression. Exp. Diabetes Res. 2012, 2012, 896362. [Google Scholar] [CrossRef] [Green Version]
- Garavelli, S.; Bruzzaniti, S.; Tagliabue, E.; Di Silvestre, D.; Prattichizzo, F.; Mozzillo, E.; Fattorusso, V.; La Sala, L.; Ceriello, A.; Puca, A.A.; et al. Plasma circulating miR-23~27~24 clusters correlate with the immunometabolic derangement and predict C-peptide loss in children with type 1 diabetes. Diabetologia 2020, 63, 2699–2712. [Google Scholar] [CrossRef]
- Salas-Pérez, F.; Codner, E.; Valencia, E.; Pizarro, C.; Carrasco, E.; Pérez-Bravo, F. MicroRNAs miR-21a and miR-93 are down regulated in peripheral blood mononuclear cells (PBMCs) from patients with type 1 diabetes. Immunobiology 2013, 218, 733–737. [Google Scholar] [CrossRef] [PubMed]
- Garavelli, S.; Bruzzaniti, S.; Tagliabue, E.; Prattichizzo, F.; Di Silvestre, D.; Perna, F.; La Sala, L.; Ceriello, A.; Mozzillo, E.; Fattorusso, V.; et al. Blood Co-Circulating Extracellular microRNAs and Immune Cell Subsets Associate with Type 1 Diabetes Severity. Int. J. Mol. Sci. 2020, 21, 477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Margaritis, K.; Margioula-Siarkou, G.; Giza, S.; Kotanidou, E.P.; Tsinopoulou, V.R.; Christoforidis, A.; Galli-Tsinopoulou, A. Micro-RNA Implications in Type-1 Diabetes Mellitus: A Review of Literature. Int. J. Mol. Sci. 2021, 22, 12165. [Google Scholar] [CrossRef]
- Aref, S.; Azmy, E.; El-Gilany, A.H. Upregulation of CD200 is associated with regulatory T cell expansion and disease progression in multiple myeloma. Hematol. Oncol. 2015, 35, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Jima, D.D.; Jacobs, C.; Fischer, R.; Gottwein, E.; Huang, G.; Lugar, P.L.; Lagoo, A.S.; Rizzieri, D.A.; Friedman, D.R.; et al. Patterns of microRNA expression characterize stages of human B-cell differentiation. Blood 2009, 113, 4586–4594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Infante, M.; Alejandro, R.; Fabbri, A.; Ricordi, C. The Heterogeneity of Type 1 Diabetes: From Immunopathology to Immune Intervention; Academic Press: Cambridge, MA, USA, 2022; pp. 83–104. [Google Scholar] [CrossRef]
- Battaglia, M.; Ahmed, S.; Anderson, M.S.; Atkinson, M.A.; Becker, D.; Bingley, P.J.; Bosi, E.; Brusko, T.M.; DiMeglio, L.A.; Evans-Molina, C.; et al. Introducing the Endotype Concept to Address the Challenge of Disease Heterogeneity in Type 1 Diabetes. Diabetes Care 2019, 43, 5–12. [Google Scholar] [CrossRef]
- Zheng, Y.; Wang, Z.; Zhou, Z. miRNAs: Novel regulators of autoimmunity-mediated pancreatic β-cell destruction in type 1 diabetes. Cell. Mol. Immunol. 2017, 14, 488–496. [Google Scholar] [CrossRef] [Green Version]
- Santos, A.S.; Cunha Neto, E.; Fukui, R.T.; Ferreira, L.R.P.; Silva, M.E.R. Increased Expression of Circulating microRNA 101-3p in Type 1 Diabetes Patients: New Insights Into miRNA-Regulated Pathophysiological Pathways for Type 1 Diabetes. Front. Immunol. 2019, 10, 1637. [Google Scholar] [CrossRef] [Green Version]
- Snowhite, I.; Pastori, R.; Sosenko, J.; Cayetano, S.M.; Pugliese, A. Baseline Assessment of Circulating MicroRNAs Near Diagnosis of Type 1 Diabetes Predicts Future Stimulated Insulin Secretion. Diabetes 2020, 70, 638–651. [Google Scholar] [CrossRef]
- Marchand, L.; Jalabert, A.; Meugnier, E.; Hende, K.V.D.; Fabien, N.; Nicolino, M.; Madec, A.-M.; Thivolet, C.; Rome, S. miRNA-375 a Sensor of Glucotoxicity Is Altered in the Serum of Children with Newly Diagnosed Type 1 Diabetes. J. Diabetes Res. 2016, 2016, 1869082. [Google Scholar] [CrossRef] [Green Version]
- Ventriglia, G.; Mancarella, F.; Sebastiani, G.; Cook, D.P.; Mallone, R.; Mathieu, C.; Gysemans, C.; Dotta, F. miR-409-3p is reduced in plasma and islet immune infiltrates of NOD diabetic mice and is differentially expressed in people with type 1 diabetes. Diabetologia 2019, 63, 124–136. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Gu, H.; Wang, L.; Huang, F.; Cai, J. MiR-885-3p is down-regulated in peripheral blood mononuclear cells from T1D patients and regulates the inflammatory response via targeting TLR4/NF-κB signaling. J. Gene Med. 2019, 22, e3145. [Google Scholar] [CrossRef] [PubMed]
- Zurawek, M.; Dzikiewicz-Krawczyk, A.; Izykowska, K.; Ziolkowska-Suchanek, I.; Skowronska, B.; Czainska, M.; Podralska, M.; Fichna, P.; Przybylski, G.; Fichna, M.; et al. miR-487a-3p upregulated in type 1 diabetes targets CTLA4 and FOXO3. Diabetes Res. Clin. Pract. 2018, 142, 146–153. [Google Scholar] [CrossRef] [PubMed]
- Samandari, N.; Mirza, A.H.; Nielsen, L.B.; Kaur, S.; Hougaard, P.; Fredheim, S.; Mortensen, H.B.; Pociot, F. Circulating microRNA levels predict residual beta cell function and glycaemic control in children with type 1 diabetes mellitus. Diabetologia 2016, 60, 354–363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomaidou, S.; Kracht, M.J.; van der Slik, A.; Laban, S.; de Koning, E.J.; Carlotti, F.; Hoeben, R.C.; Roep, B.O.; Zaldumbide, A. β-Cell Stress Shapes CTL Immune Recognition of Preproinsulin Signal Peptide by Posttranscriptional Regulation of Endoplasmic Reticulum Aminopeptidase 1. Diabetes 2020, 69, 670–680. [Google Scholar] [CrossRef] [PubMed]
- Tsukita, S.; Yamada, T.; Takahashi, K.; Munakata, Y.; Hosaka, S.; Takahashi, H.; Gao, J.; Shirai, Y.; Kodama, S.; Asai, Y.; et al. MicroRNAs 106b and 222 Improve Hyperglycemia in a Mouse Model of Insulin-Deficient Diabetes via Pancreatic β-Cell Proliferation. Ebiomedicine 2016, 15, 163–172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mandelbaum, A.D.; Kredo-Russo, S.; Aronowitz, D.; Myers, N.; Yanowski, E.; Klochendler, A.; Swisa, A.; Dor, Y.; Hornstein, E. miR-17-92 and miR-106b-25 clusters regulate beta cell mitotic checkpoint and insulin secretion in mice. Diabetologia 2019, 62, 1653–1666. [Google Scholar] [CrossRef]
- Xin, Y.; Cai, H.; Lu, T.; Zhang, Y.; Yang, Y.; Cui, Y. miR-20b Inhibits T Cell Proliferation and Activation via NFAT Signaling Pathway in Thymoma-Associated Myasthenia Gravis. BioMed Res. Int. 2016, 2016, 9595718. [Google Scholar] [CrossRef] [Green Version]
- Ji, H.; Fan, L.; Shan, A.; Wang, W.; Ning, G.; Cao, Y.; Jiang, X. Let7b-5p inhibits insulin secretion and decreases pancreatic β-cell mass in mice. Mol. Cell. Endocrinol. 2022, 540. [Google Scholar] [CrossRef]
- Sung, Y.; Jeong, J.; Kang, R.J.; Choi, M.; Park, S.; Kwon, W.; Lee, J.; Jang, S.; Park, S.J.; Kim, S.; et al. Lin28a expression protects against streptozotocin-induced β-cell destruction and prevents diabetes in mice. Cell Biochem. Funct. 2019, 37, 139–147. [Google Scholar] [CrossRef]
- Zurawek, M.; Fichna, M.; Fichna, P.; Czainska, M.; Rozwadowska, N. Upregulation of FOXO3 in New-Onset Type 1 Diabetes Mellitus. J. Immunol. Res. 2020, 2020, 9484015. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Muniappan, L.; Tang, G.; Özcan, S. Identification of glucose-regulated miRNAs from pancreatic β cells reveals a role for miR-30d in insulin transcription. Rna 2008, 15, 287–293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, X.; Mohan, R.; Özcan, S.; Tang, X. MicroRNA-30d Induces Insulin Transcription Factor MafA and Insulin Production by Targeting Mitogen-activated Protein 4 Kinase 4 (MAP4K4) in Pancreatic β-Cells. J. Biol. Chem. 2012, 287, 31155–31164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaziel-Sovran, A.; Segura, M.F.; Di Micco, R.; Collins, M.K.; Hanniford, D.; de Miera, E.V.-S.; Rakus, J.F.; Dankert, J.F.; Shang, S.; Kerbel, R.S.; et al. miR-30b/30d Regulation of GalNAc Transferases Enhances Invasion and Immunosuppression during Metastasis. Cancer Cell 2011, 20, 104–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, S.; Rezende, R.M.; Moreira, T.G.; Tankou, S.K.; Cox, L.M.; Wu, M.; Song, A.; Dhang, F.H.; Wei, Z.; Costamagna, G.; et al. Oral Administration of miR-30d from Feces of MS Patients Suppresses MS-like Symptoms in Mice by Expanding Akkermansia muciniphila. Cell Host Microbe 2019, 26, 779–794.e8. [Google Scholar] [CrossRef] [PubMed]
- Seyhan, A.A.; Lopez, Y.O.N.; Xie, H.; Yi, F.; Mathews, C.; Pasarica, M.; Pratley, R.E. Pancreas-enriched miRNAs are altered in the circulation of subjects with diabetes: A pilot cross-sectional study. Sci. Rep. 2016, 6, 31479. [Google Scholar] [CrossRef] [Green Version]
- Frørup, C.; Mirza, A.H.; Yarani, R.; Nielsen, L.B.; Mathiesen, E.R.; Damm, P.; Svare, J.; Engelbrekt, C.; Størling, J.; Johannesen, J.; et al. Plasma Exosome-Enriched Extracellular Vesicles From Lactating Mothers With Type 1 Diabetes Contain Aberrant Levels of miRNAs During the Postpartum Period. Front. Immunol. 2021, 12, 744509. [Google Scholar] [CrossRef]
- Qian, X.; Yang, Z.; Mao, E.; Chen, E. Regulation of fatty acid synthesis in immune cells. Scand. J. Immunol. 2018, 88, e12713. [Google Scholar] [CrossRef] [Green Version]
- Coles, S.J.; Hills, R.K.; Wang, E.C.Y.; Burnett, A.K.; Man, S.; Darley, R.L.; Tonks, A. Increased CD200 expression in acute myeloid leukemia is linked with an increased frequency of FoxP3+ regulatory T cells. Leukemia 2012, 26, 2146–2148. [Google Scholar] [CrossRef]
- Gorczynski, R.; Chen, Z.; Khatri, I.; Yu, K. sCD200 Present in Mice Receiving Cardiac and Skin Allografts Causes Immunosuppression In Vitro and Induces Tregs. Transplantation 2013, 95, 442–447. [Google Scholar] [CrossRef]
- Sanda, S.; Roep, B.O.; von Herrath, M. Islet antigen specific IL-10+ immune responses but not CD4+CD25+FoxP3+ cells at diagnosis predict glycemic control in type 1 diabetes. Clin. Immunol. 2008, 127, 138–143. [Google Scholar] [CrossRef] [PubMed]
- Fauth, M.; Hegewald, A.B.; Schmitz, L.; Krone, D.J.; Saul, M.J. Validation of extracellular miRNA quantification in blood samples using RT-qPCR. FASEB BioAdvances 2019, 1, 481–492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elmén, J.; Lindow, M.; Schütz, S.; Lawrence, M.; Petri, A.; Obad, S.; Lindholm, M.; Hedtjärn, M.; Hansen, H.F.; Berger, U.; et al. LNA-mediated microRNA silencing in non-human primates. Nature 2008, 452, 896–899. [Google Scholar] [CrossRef] [PubMed]
- Serr, I.; Scherm, M.G.; Zahm, A.M.; Schug, J.; Flynn, V.K.; Hippich, M.; Kälin, S.; Becker, M.; Achenbach, P.; Nikolaev, A.; et al. A miRNA181a/NFAT5 axis links impaired T cell tolerance induction with autoimmune type 1 diabetes. Sci. Transl. Med. 2018, 10, eaag1782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krützfeldt, J.; Rajewsky, N.; Braich, R.; Rajeev, K.G.; Tuschl, T.; Manoharan, M.; Stoffel, M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 2005, 438, 685–689. [Google Scholar] [CrossRef] [PubMed]
- Elmén, J.; Lindow, M.; Silahtaroglu, A.; Bak, M.; Christensen, M.; Lind-Thomsen, A.; Hedtjärn, M.; Hansen, J.B.; Hansen, H.F.; Straarup, E.M.; et al. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 2007, 36, 1153–1162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- American Diabetes Association. Classification and Diagnosis of Diabetes: Standards of Medical Care in Diabetes—2021. Diabetes Care 2021, 44, S15–S33. [Google Scholar] [CrossRef]
- Lezcano, A.C.; García, J.F.; Ramos, C.F.; Longás, A.F.; López-Siguero, J.; González, E.S.; Ruiz, B.S.; Fernández, D.Y. Estudio transversal español de crecimiento 2008. Parte II: Valores de talla, peso e índice de masa corporal desde el nacimiento a la talla adulta. In Anales de Pediatría; Elsevier Doyma: Hoboken, NJ, USA, 2008; Volume 68, pp. 552–569. [Google Scholar] [CrossRef]
- Mortensen, H.B.; Hougaard, P.; Swift, P.; Hansen, L.; Holl, R.W.; Hoey, H.; Bjoerndalen, H.; de Beaufort, C.; Chiarelli, F.; Danne, T.; et al. New Definition for the Partial Remission Period in Children and Adolescents With Type 1 Diabetes. Diabetes Care 2009, 32, 1384–1390. [Google Scholar] [CrossRef] [Green Version]
- Vlachos, I.S.; Zagganas, K.; Paraskevopoulou, M.D.; Georgakilas, G.; Karagkouni, D.; Vergoulis, T.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-miRPath v3.0: Deciphering microRNA function with experimental support. Nucleic Acids Res. 2015, 43, W460–W466. [Google Scholar] [CrossRef]
- Vlachos, I.S.; Paraskevopoulou, M.D.; Karagkouni, D.; Georgakilas, G.; Vergoulis, T.; Kanellos, I.; Anastasopoulos, I.-L.; Maniou, S.; Karathanou, K.; Kalfakakou, D.; et al. DIANA-TarBase v7.0: Indexing more than half a million experimentally supported miRNA:mRNA interactions. Nucleic Acids Res. 2014, 43, D153–D159. [Google Scholar] [CrossRef]
- Supek, F.; Bošnjak, M.; Škunca, N.; Smuc, T. REVIGO Summarizes and Visualizes Long Lists of Gene Ontology Terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, M.D.; Leemaqz, S.Y.; Jankovic-Karasoulos, T.; McAninch, D.; McCullough, D.; Breen, J.; Roberts, C.T.; Pillman, K.A. Haemolysis Detection in MicroRNA-Seq from Clinical Plasma Samples. Genes 2022, 13, 1288. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of Real-Time Quantitative Reverse Transcription-PCR Data: A Model-Based Variance Estimation Approach to Identify Genes Suited for Normalization, Applied to Bladder and Colon Cancer Data Sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [Green Version]
- Alba, A.; Puertas, M.C.; Carrillo, J.; Planas, R.; Ampudia, R.; Pastor, X.; Bosch, F.; Pujol-Borrell, R.; Verdaguer, J.; Vives-Pi, M. IFNβ Accelerates Autoimmune Type 1 Diabetes in Nonobese Diabetic Mice and Breaks the Tolerance to β Cells in Nondiabetes-Prone Mice. J. Immunol. 2004, 173, 6667–6675. [Google Scholar] [CrossRef] [PubMed] [Green Version]









| Discovery Cohort | Validation Cohort | |||||||
|---|---|---|---|---|---|---|---|---|
| Ctrl | T1D dx | PR | Non-PR | Ctrl | T1D dx | PR | Non-PR | |
| N (no. girls) | 17 (10) | 17 (10) | 11 (6) | 6 (4) | 15 (9) | 8 (4) | 10 (5) | 9 (7) |
| Age at diagnosis (years) | 8.8 ± 3.4 | 8.7 ± 3.6 | 9.1 ± 4.3 | 9 ± 2.8 | 9.7 ± 3.6 | 11.6 ± 2.7 | 11.8 ± 2.9 | 7.3 ± 3.7 |
| BMI (kg/m2) | 18.3 ± 4.3 | 16.8 ± 2.5 | 17.7 ± 3 | 17.2 ± 2.1 | 19 ± 4.2 | 19.4 ± 4.3 | 20.2 ± 4.6 | 16.9 ± 1.7 |
| HbA1c (%) | ND | 11.4 ± 2.4 | 6.9 ± 0.6 | 8.1 ± 0.7 | ND | 12.6 ± 2.5 | 6.4 ± 0.6 | 8.3 ± 0.6 |
| Insulin dose (U/kg/day) | ND | 0.7 ± 0.2 | 0.4 ± 0.1 | 0.9 ± 0.1 | ND | 0.77 ± 0.2 | 0.46 ± 0.2 | 0.87 ± 0.2 |
| IDAA1c | ND | 14.3 ± 3 | 8.4 ± 0.5 | 11.5 ± 1 | ND | 15.7 ± 3.1 | 8.2 ± 0.79 | 11.8 ± 1.4 |
| Basal C-peptide (ng/mL) | 1.3 ± 0.4 | 0.3 ± 0.2 | 0.7 ± 0.5 | 0.3 ± 0.1 | 1.97 ± 0.4 | 0.44 ± 0.3 | 1 ± 0.57 | 0.2 ± 0.1 |
| miRNAs | PR vs. Ctrl (FC) | p-Value | PR vs. T1D dx (FC) | p-Value | PR vs. Non-PR (FC) | p-Value |
|---|---|---|---|---|---|---|
| hsa-miR-30d-5p | 1.777 | 0.019 | - | - | 3.208 | 0.001 |
| hsa-let-7b-5p | - | - | - | - | −1.019 | 0.005 |
| hsa-miR-106a-5p | - | - | - | - | 2.771 | 0.006 |
| hsa-miR-20b-5p | - | - | - | - | 2.842 | 0.006 |
| hsa-let-7c-5p | - | - | - | - | −2.587 | 0.009 |
| hsa-miR-25-5p | −1.921 | 0.019 | - | - | −2.722 | 0.014 |
| hsa-miR-320b | - | - | - | - | 2.477 | 0.020 |
| hsa-miR-30e-5p | - | - | - | - | 2.485 | 0.022 |
| hsa-miR-142-3p | - | - | - | - | 1.125 | 0.023 |
| hsa-miR-106b-5p | - | - | - | - | 2.361 | 0.030 |
| hsa-miR-144-3p | - | - | −1.027 | 0.006 | 1.064 | 0.032 |
| hsa-miR-18b-5p | - | - | - | - | 2.235 | 0.043 |
| hsa-miR-17-5p | - | - | - | - | 1.319 | 0.048 |
| hsa-miR-1277-3p | 2.126 | 0.008 | - | - | 2.162 | 0.049 |
| hsa-miR-101-5p | −1.935 | 0.015 | −2.910 | 0.002 | - | - |
| hsa-miR-4485-3p | - | - | −2.890 | 0.005 | - | - |
| hsa-miR-10b-5p | - | - | 2.697 | 0.005 | - | - |
| hsa-miR-1976 | −1.568 | 0.037 | −2.433 | 0.008 | - | - |
| hsa-miR-296-5p | −1.830 | 0.018 | −2.444 | 0.009 | - | - |
| hsa-miR-377-3p | −2.028 | 0.016 | −2.558 | 0.013 | - | - |
| hsa-miR-543 | −1.838 | 0.023 | −2.311 | 0.018 | - | - |
| hsa-miR-24-3p | - | - | 2.069 | 0.019 | - | - |
| hsa-miR-3611 | −2.293 | 0.002 | −2.108 | 0.019 | - | - |
| hsa-miR-4485-5p | −-2.883 | 0.005 | −2.846 | 0.020 | - | - |
| hsa-miR-154-5p | −2.556 | 0.001 | −2.256 | 0.020 | - | - |
| hsa-miR-223-3p | - | - | 1.311 | 0.021 | - | - |
| hsa-miR-324-3p | - | - | −2.244 | 0.022 | - | - |
| hsa-miR-1-3p | - | - | −2.263 | 0.029 | - | - |
| hsa-miR-4449 | - | - | −1.951 | 0.037 | - | - |
| hsa-miR-365a-3p | - | - | 2.059 | 0.039 | - | - |
| hsa-miR-365b-3p | - | - | 2.059 | 0.039 | - | - |
| hsa-miR-4446-3p | −2.194 | 0.009 | −2.025 | 0.043 | - | - |
| hsa-miR-132-3p | - | - | −1.943 | 0.045 | - | - |
| miRNA | Assay ID |
|---|---|
| hsa-miR-142-3p | 477910_mir |
| hsa-miR-20b-5p | 477804_mir |
| hsa-miR-17-5p | 478447_mir |
| hsa-let-7b-5p | 478576_mir |
| hsa-let-7c-5p | 478577_mir |
| hsa-miR-106b-5p | 478412_mir |
| ath-miR159a | 478411_mir |
| hsa-miR-16-1-3p | 478727_mir |
| Target | Fluorophore | Species/Isotype | Clone | Use/100 µL | Company | |
|---|---|---|---|---|---|---|
| PANEL 1 | CD3 | PE | Hamster IgG | 500A2 | 1 | BD Biosciences |
| CD4 | APC | Rat IgG2a, κ | RM4-5 | 0.5 | BD Biosciences | |
| CD8 | V500 | Rat IgG2a, κ | 53-6.7 | 0.4 | BD Biosciences | |
| CD44 | BV786 | Rat IgG2b, κ | IM7 | 0.2 | BD Biosciences | |
| CD62L | APC-Cy7 | Rat IgG2a, κ | MEL-14 | 0.4 | BioLegend | |
| PD-1 | PE-Cy7 | Rat IgG2a, κ | 29F.1A12 | 2 | BioLegend | |
| CD25 | PerCP-Cy5.5 | Rat/IgG1, λ | PC61.5 | 1 | eBioscience | |
| FOXP3 | FITC | Rat IgG2a, κ | FJK-16s | 1.5 | eBioscience | |
| PANEL 2 | CD3 | PE | Hamster IgG | 500A2 | 1 | BD Biosciences |
| CD19 | V450 | Rat IgG2a, κ | 1D3 | 1 | BD Biosciences | |
| CD11c | PE-Cy7 | Hamster IgG1, λ2 | HL3 | 1 | BD Biosciences | |
| MHC-II | APC | Rat IgG2b, κ | AMS-32.1 | 1 | eBioscience |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gomez-Muñoz, L.; Perna-Barrull, D.; Murillo, M.; Armengol, M.P.; Alcalde, M.; Catala, M.; Rodriguez-Fernandez, S.; Sunye, S.; Valls, A.; Perez, J.; et al. Immunoregulatory Biomarkers of the Remission Phase in Type 1 Diabetes: miR-30d-5p Modulates PD-1 Expression and Regulatory T Cell Expansion. Non-Coding RNA 2023, 9, 17. https://doi.org/10.3390/ncrna9020017
Gomez-Muñoz L, Perna-Barrull D, Murillo M, Armengol MP, Alcalde M, Catala M, Rodriguez-Fernandez S, Sunye S, Valls A, Perez J, et al. Immunoregulatory Biomarkers of the Remission Phase in Type 1 Diabetes: miR-30d-5p Modulates PD-1 Expression and Regulatory T Cell Expansion. Non-Coding RNA. 2023; 9(2):17. https://doi.org/10.3390/ncrna9020017
Chicago/Turabian StyleGomez-Muñoz, Laia, David Perna-Barrull, Marta Murillo, Maria Pilar Armengol, Marta Alcalde, Marti Catala, Silvia Rodriguez-Fernandez, Sergi Sunye, Aina Valls, Jacobo Perez, and et al. 2023. "Immunoregulatory Biomarkers of the Remission Phase in Type 1 Diabetes: miR-30d-5p Modulates PD-1 Expression and Regulatory T Cell Expansion" Non-Coding RNA 9, no. 2: 17. https://doi.org/10.3390/ncrna9020017