3D Co-Printing and Substrate Geometry Influence the Differentiation of C2C12 Skeletal Myoblasts
Abstract
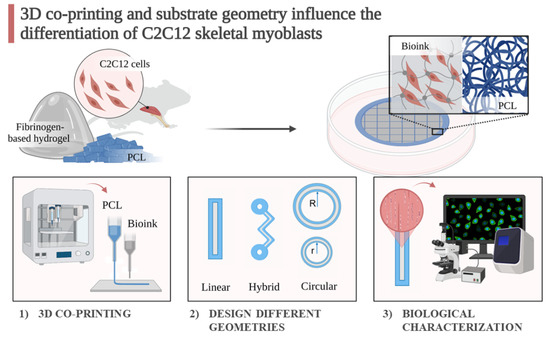
:1. Introduction
2. Results and Discussion
2.1. Linear Structures: Bioprinted vs. Co-Printed
2.1.1. Gene Expression Analysis by Quantitative Real-Time PCR
2.2. Curvature Feature: Circular and Hybrid Structures
Gene Expression Analysis by Quantitative Real-Time PCR
2.3. Comparison of All Tested Geometries
2.3.1. Live/Dead Staining
2.3.2. Gene Expression Analysis by Quantitative Real-Time PCR
2.4. Discussion
2.5. Limitations
3. Conclusions
4. Materials and Methods
4.1. Cell Culture
4.2. Biomaterials and Crosslinker
4.3. 3D Bioprinter
4.4. Co-Printing Process and 3D Constructs Culture
4.5. 3D Structure
4.6. Live/Dead Staining
4.7. Total RNA Extraction and Quantitative Real-Time PCR
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, C.S.; Tan, J.; Tien, J. Mechanotransduction at Cell-Matrix and Cell-Cell Contacts. Annu. Rev. Biomed. Eng. 2004, 6, 275–302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bajaj, P.; Tang, X.; Saif, T.A.; Bashir, R. Stiffness of the substrate influences the phenotype of embryonic chicken cardiac myocytes. J. Biomed. Mater. Res. A 2010, 95, 1261–1269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Discher, D.E.; Janmey, P.; Wang, Y.-L. Tissue Cells Feel and Respond to the Stiffness of Their Substrate. Science 2005, 310, 1139–1143. [Google Scholar] [CrossRef] [Green Version]
- Engler, A.J.; Griffin, M.A.; Sen, S.; Bonnemann, C.G.; Sweeney, H.L.; Discher, D.E. Myotubes differentiate optimally on substrates with tissue-like stiffness: Pathological implications for soft or stiff microenvironments. J. Cell Biol. 2004, 166, 877–887. [Google Scholar] [CrossRef]
- Solon, J.; Levental, I.; Sengupta, K.; Georges, P.; Janmey, P.A. Fibroblast Adaptation and Stiffness Matching to Soft Elastic Substrates. Biophys. J. 2007, 93, 4453–4461. [Google Scholar] [CrossRef] [Green Version]
- Yeung, T.; Georges, P.C.; Flanagan, L.A.; Marg, B.; Ortiz, M.; Funaki, M.; Zahir, N.; Ming, W.; Weaver, V.; Janmey, P.A. Effects of substrate stiffness on cell morphology, cytoskeletal structure, and adhesion. Cell Motil. Cytoskelet. 2005, 60, 24–34. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Dean, D.; Wallace, J.; Breithaupt, R.; Mikos, A.G.; Fisher, J.P. The influence of stereolithographic scaffold architecture and composition on osteogenic signal expression with rat bone marrow stromal cells. Biomaterials 2011, 32, 3750–3763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.S.; Mrksich, M.; Huang, S.; Whitesides, G.M.; Ingber, D.E. Geometric Control of Cell Life and Death. Science 1997, 276, 1425–1428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balaban, N.Q.; Schwarz, U.; Riveline, D.; Goichberg, P.; Tzur, G.; Sabanay, I.; Mahalu, D.; Safran, S.; Bershadsky, A.; Addadiand, L.; et al. Force and focal adhesion assembly: A close relationship studied using elastic micropatterned substrates. Nat. Cell Biol. 2001, 3, 466–472. [Google Scholar] [CrossRef]
- Huang, N.F.; Patel, S.; Thakar, R.G.; Wu, J.; Hsiao, B.S.; Chu, B.; Lee, R.J.; Li, S. Myotube Assembly on Nanofibrous and Micropatterned Polymers. Nano Lett. 2006, 6, 537–542. [Google Scholar] [CrossRef]
- Lee, L.H.; Peerani, R.; Ungrin, M.; Joshi, C.; Kumacheva, E.; Zandstra, P. Micropatterning of human embryonic stem cells dissects the mesoderm and endoderm lineages. Stem Cell Res. 2009, 2, 155–162. [Google Scholar] [CrossRef] [Green Version]
- Yao, Y.-T.; Yang, Y.; Ye, Q.; Cao, S.-S.; Zhang, X.-P.; Zhao, K.; Jian, Y. Effects of pore size and porosity on cytocompatibility and osteogenic differentiation of porous titanium. J. Mater. Sci. Mater. Med. 2021, 32, 72. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Lian, M.; Wu, Q.; Qiao, Z.; Sun, B.; Dai, K. Effect of Pore Size on Cell Behavior Using Melt Electrowritten Scaffolds. Front. Bioeng. Biotechnol. 2021, 9, 629270. [Google Scholar] [CrossRef]
- Kilian, K.A.; Bugarija, B.; Lahn, B.T.; Mrksich, M. Geometric cues for directing the differentiation of mesenchymal stem cells. Proc. Natl. Acad. Sci. USA 2010, 107, 4872–4877. [Google Scholar] [CrossRef] [PubMed]
- Bajaj, P.; Reddy, B.; Millet, L.; Wei, C.; Zorlutuna, P.; Bao, G.; Bashir, R. Patterning the differentiation of C2C12 skeletal myoblasts. Integr. Biol. 2011, 3, 897–909. [Google Scholar] [CrossRef] [PubMed]
- Vogel, V.; Sheetz, M. Local force and geometry sensing regulate cell functions. Nat. Rev. Mol. Cell Biol. 2006, 7, 265–275. [Google Scholar] [CrossRef]
- Bahcecioglu, G.; Basara, G.; Ellis, B.W.; Ren, X.; Zorlutuna, P. Breast cancer models: Engineering the tumor microenvironment. Acta Biomater. 2020, 106, 1–21. [Google Scholar] [CrossRef]
- Malda, J.; Visser, J.; Melchels, F.P.; Jüngst, T.; Hennink, W.E.; Dhert, W.J.A.; Groll, J.; Hutmacher, D.W. 25th Anniversary Article: Engineering Hydrogels for Biofabrication. Adv. Mater. 2013, 25, 5011–5028. [Google Scholar] [CrossRef]
- Ronzoni, F.L.; Aliberti, F.; Scocozza, F.; Benedetti, L.; Auricchio, F.; Sampaolesi, M.; Cusella, G.; Redwan, I.N.; Ceccarelli, G.; Conti, M. Myoblast 3D bioprinting to burst in vitro skeletal muscle differentiation. J. Tissue Eng. Regen. Med. 2022, 16, 484–495. [Google Scholar] [CrossRef]
- Bade, N.D.; Kamien, R.D.; Assoian, R.K.; Stebe, K.J. Curvature and Rho activation differentially control the alignment of cells and stress fibers. Sci. Adv. 2017, 3, e1700150. [Google Scholar] [CrossRef] [Green Version]
- Rumpler, M.; Woesz, A.; Dunlop, J.W.; van Dongen, J.T.; Fratzl, P. The effect of geometry on three-dimensional tissue growth. J. R. Soc. Interface 2008, 5, 1173–1180. [Google Scholar] [CrossRef] [Green Version]
- Bidan, C.M.; Kommareddy, K.P.; Rumpler, M.; Kollmannsberger, P.; Bréchet, Y.J.M.; Fratzl, P.; Dunlop, J.W.C. How Linear Tension Converts to Curvature: Geometric Control of Bone Tissue Growth. PLoS ONE 2012, 7, e36336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bidan, C.M.; Kommareddy, K.P.; Rumpler, M.; Kollmannsberger, P.; Fratzl, P.; Dunlop, J.W.C. Geometry as a Factor for Tissue Growth: Towards Shape Optimization of Tissue Engineering Scaffolds. Adv. Healthc. Mater. 2013, 2, 186–194. [Google Scholar] [CrossRef] [PubMed]
- Knychala, J.; Bouropoulos, N.; Catt, C.J.; Katsamenis, O.L.; Please, C.P.; Sengers, B.G. Pore Geometry Regulates Early Stage Human Bone Marrow Cell Tissue Formation and Organisation. Ann. Biomed. Eng. 2013, 41, 917–930. [Google Scholar] [CrossRef] [PubMed]
- Guyot, Y.; Papantoniou, I.; Chai, Y.C.; Van Bael, S.; Schrooten, J.; Geris, L. A computational model for cell/ECM growth on 3D surfaces using the level set method: A bone tissue engineering case study. Biomech. Model. Mechanobiol. 2014, 13, 1361–1371. [Google Scholar] [CrossRef]
- Guyot, Y.; Papantoniou, I.; Luyten, F.P.; Geris, L. Coupling curvature-dependent and shear stress-stimulated neotissue growth in dynamic bioreactor cultures: A 3D computational model of a complete scaffold. Biomech. Model. Mechanobiol. 2016, 15, 169–180. [Google Scholar] [CrossRef] [Green Version]
- Werner, M.; Kurniawan, N.A.; Korus, G.; Bouten, C.V.C.; Petersen, A. Mesoscale substrate curvature overrules nanoscale contact guidance to direct bone marrow stromal cell migration. J. R. Soc. Interface 2018, 15, 20180162. [Google Scholar] [CrossRef] [Green Version]
- Callens, S.J.; Uyttendaele, R.J.; Fratila-Apachitei, L.E.; Zadpoor, A.A. Substrate curvature as a cue to guide spatiotemporal cell and tissue organization. Biomaterials 2019, 232, 119739. [Google Scholar] [CrossRef]
- Huang, Y.-C.; Dennis, R.G.; Larkin, L.; Baar, K. Rapid formation of functional muscle in vitro using fibrin gels. J. Appl. Physiol. 2005, 98, 706–713. [Google Scholar] [CrossRef] [Green Version]
- Bari, E.; Scocozza, F.; Perteghella, S.; Sorlini, M.; Auricchio, F.; Torre, M.L.; Conti, M. 3D Bioprinted Scaffolds Containing Mesenchymal Stem/Stromal Lyosecretome: Next Generation Controlled Release Device for Bone Regenerative Medicine. Pharmaceutics 2021, 13, 515. [Google Scholar] [CrossRef]
- Bari, E.; Scocozza, F.; Perteghella, S.; Segale, L.; Sorlini, M.; Auricchio, F.; Conti, M.; Torre, M.L. Three-Dimensional Bioprinted Controlled Release Scaffold Containing Mesenchymal Stem/Stromal Lyosecretome for Bone Regeneration: Sterile Manufacturing and In Vitro Biological Efficacy. Biomedicines 2022, 10, 1063. [Google Scholar] [CrossRef]
- Izadifar, Z.; Chang, T.; Kulyk, W.; Chen, X.; Eames, B.F. Analyzing Biological Performance of 3D-Printed, Cell-Impregnated Hybrid Constructs for Cartilage Tissue Engineering. Tissue Eng. Part C Methods 2016, 22, 173–188. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.S.; Kim, G. Three-dimensional electrospun polycaprolactone (PCL)/alginate hybrid composite scaffolds. Carbohydr. Polym. 2014, 114, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.B.; Kim, G.H. PCL/Alginate Composite Scaffolds for Hard Tissue Engineering: Fabrication, Characterization, and Cellular Activities. ACS Comb. Sci. 2015, 17, 87–99. [Google Scholar] [CrossRef]
- Merceron, T.K.; Burt, M.; Seol, Y.-J.; Kang, H.-W.; Lee, S.J.; Yoo, J.J.; Atala, A. A 3D bioprinted complex structure for engineering the muscle–tendon unit. Biofabrication 2015, 7, 035003. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.-J.; Kim, T.G.; Jeong, J.; Yi, H.-G.; Park, J.W.; Hwang, W.; Cho, D.-W. 3D Cell Printing of Functional Skeletal Muscle Constructs Using Skeletal Muscle-Derived Bioink. Adv. Health Mater. 2016, 5, 2636–2645. [Google Scholar] [CrossRef]
- Kang, H.-W.; Lee, S.J.; Ko, I.K.; Kengla, C.; Yoo, J.J.; Atala, A. A 3D bioprinting system to produce human-scale tissue constructs with structural integrity. Nat. Biotechnol. 2016, 34, 312–319. [Google Scholar] [CrossRef]
- Kim, J.H.; Seol, Y.-J.; Ko, I.K.; Kang, H.-W.; Lee, Y.K.; Yoo, J.J.; Atala, A.; Lee, S.J. 3D Bioprinted Human Skeletal Muscle Constructs for Muscle Function Restoration. Sci. Rep. 2018, 8, 12307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Callens, S.J.P.; Fan, D.; van Hengel, I.A.J.; Minneboo, M.; Díaz-Payno, P.J.; Stevens, M.M.; Fratila-Apachitei, L.E.; Zadpoor, A.A. Emergent collective organization of bone cells in complex curvature fields. Nat. Commun. 2023, 14, 855. [Google Scholar] [CrossRef]
- Wang, P.-Y.; Yu, H.-T.; Tsai, W.-B. Modulation of alignment and differentiation of skeletal myoblasts by submicron ridges/grooves surface structure. Biotechnol. Bioeng. 2010, 106, 285–294. [Google Scholar] [CrossRef]
- Shimizu, K.; Fujita, H.; Nagamori, E. Alignment of skeletal muscle myoblasts and myotubes using linear micropatterned surfaces ground with abrasives. Biotechnol. Bioeng. 2009, 103, 631–638. [Google Scholar] [CrossRef]
- Yamamoto, D.L.; Csikasz, R.I.; Li, Y.; Sharma, G.; Hjort, K.; Karlsson, R.; Bengtsson, T. Myotube Formation on Micro-patterned Glass: Intracellular Organization and Protein Distribution in C2C12 Skeletal Muscle Cells. J. Histochem. Cytochem. 2008, 56, 881–892. [Google Scholar] [CrossRef] [Green Version]
- Feinberg, A.W.; Feigel, A.; Shevkoplyas, S.S.; Sheehy, S.; Whitesides, G.M.; Parker, K.K. Muscular Thin Films for Building Actuators and Powering Devices. Science 2007, 317, 1366–1370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanaka, Y.; Morishima, K.; Shimizu, T.; Kikuchi, A.; Yamato, M.; Okano, T.; Kitamori, T. An actuated pump on-chip powered by cultured cardiomyocytes. Lab Chip 2006, 6, 362–368. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Morishima, K.; Shimizu, T.; Kikuchi, A.; Yamato, M.; Okano, T.; Kitamori, T. Demonstration of a PDMS-based bio-microactuator using cultured cardiomyocytes to drive polymer micropillars. Lab Chip 2006, 6, 230–235. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, Y.; Furukawa, Y.; Morishima, K. Controllable Bio-Microactuator Powered by Muscle Cells. In Proceedings of the 2006 International Conference of the IEEE Engineering in Medicine and Biology Society, New York, NY, USA, 5 March 2008; pp. 6565–6568. [Google Scholar] [CrossRef]
- Herr, H.; Dennis, R.G. A swimming robot actuated by living muscle tissue. J. Neuroeng. Rehabil. 2004, 1, 6. [Google Scholar] [CrossRef] [Green Version]
- Rao, X.; Huang, X.; Zhou, Z.; Lin, X. An improvement of the 2ˆ(–delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat. Bioinform. Biomath. 2013, 3, 71–85. [Google Scholar]





| Operational Parameters | PCL | CELLINK FIBRIN |
|---|---|---|
| Extrusion pressure [kPa] | 300 | 10–15 |
| Conical nozzle diameter [mm] | 0.5 | 0.41 |
| Printing speed [mm/min] | 45 | 600 |
| Printing temperature [°C] | 90 | RT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Loi, G.; Scocozza, F.; Aliberti, F.; Rinvenuto, L.; Cidonio, G.; Marchesi, N.; Benedetti, L.; Ceccarelli, G.; Conti, M. 3D Co-Printing and Substrate Geometry Influence the Differentiation of C2C12 Skeletal Myoblasts. Gels 2023, 9, 595. https://doi.org/10.3390/gels9070595
Loi G, Scocozza F, Aliberti F, Rinvenuto L, Cidonio G, Marchesi N, Benedetti L, Ceccarelli G, Conti M. 3D Co-Printing and Substrate Geometry Influence the Differentiation of C2C12 Skeletal Myoblasts. Gels. 2023; 9(7):595. https://doi.org/10.3390/gels9070595
Chicago/Turabian StyleLoi, Giada, Franca Scocozza, Flaminia Aliberti, Lorenza Rinvenuto, Gianluca Cidonio, Nicola Marchesi, Laura Benedetti, Gabriele Ceccarelli, and Michele Conti. 2023. "3D Co-Printing and Substrate Geometry Influence the Differentiation of C2C12 Skeletal Myoblasts" Gels 9, no. 7: 595. https://doi.org/10.3390/gels9070595