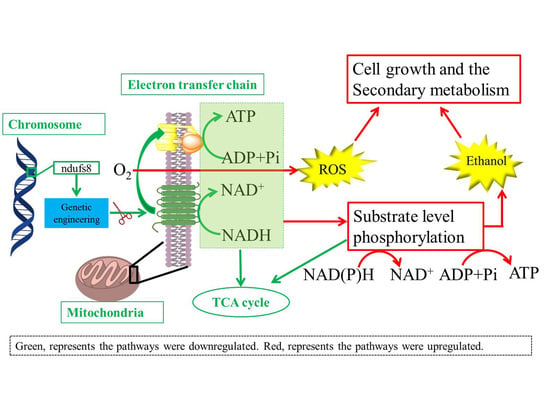
Role of the Gene ndufs8 Located in Respiratory Complex I from Monascus purpureus in the Cell Growth and Secondary Metabolites Biosynthesis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microorganisms and Culture Conditions
2.2. Sensitivity Test
2.3. Image Analysis
2.4. Submerged Batch-Fermentation and Kinetics
2.5. Enzyme Activity Detection
2.6. Determination of Metabolites and Calculations
2.7. Transcriptomic Analysis
2.8. Validation of Gene Expression Levels via RT-qPCR
2.9. Data Analysis
2.10. Accession Numbers
3. Results and Discussion
3.1. Functional Annotation of Gene monascus_4971
3.2. Determination of the Transformant Strain
3.3. Effect of ndufs8 Gene Located in Mitochondrial Complex I on Monascus Growth and Morphology
3.4. Effect of ndufs8 Gene on the Monascus Secondary Metabolism
3.5. Regulation Mechanisms of ndufs8 Gene in Monascus
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xiong, Z.; Cao, X.; Wen, Q.; Chen, Z.; Huang, Z. An overview of the bioactivity of monacolin K/lovastatin. Food Chem. Toxicol. 2019, 131, 110585. [Google Scholar] [CrossRef] [PubMed]
- Patakova, P.; Branska, B.; Patrovsky, M. Monascus Secondary Metabolites. In Fungal Metabolites; Reference Series in Phytochemistry; Mérillon, J.M., Ramawat, K., Eds.; Springer: Cham, Switzerland, 2017; pp. 821–851. [Google Scholar] [CrossRef]
- Dufossé, L.; Galaup, P.; Yaron, A.; Arad, S.M.; Blanc, P.; Chidambara Murthy, K.N.; Ravishankar, G.A. Microorganisms and microalgae as sources of pigments for food use: A scientific oddity or an industrial reality? Trends Food Sci. Technol. 2005, 16, 389–406. [Google Scholar] [CrossRef]
- Xiao, Q.J.; Zhi, N.X.; Li, P.Z.; Chang, K.S. Elimination of the mycotoxin citrinin production in the industrial important strain Monascus purpureus SM001. Metab. Eng. 2010, 12, 1–7. [Google Scholar]
- Blanc, P.J.; Laussac, J.P.; Bars, J.L.; Bars, P.L.; Loret, M.O.; Pareilleux, A.; Prome, D.; Prome, J.C.; Santerre, A.L.; Goma, G. Characterization of monascidin A from Monascus as citrinin. Int. J. Food Microbiol. 1995, 27, 201–213. [Google Scholar] [CrossRef]
- Chen, W.; Chen, R.; Liu, Q.; He, Y.; He, K.; Ding, X.; Kang, L.; Guo, X.; Xie, N.; Zhou, Y.; et al. Orange, red, yellow: Biosynthesis of azaphilone pigments in Monascus fungi. Chem. Sci. 2017, 7, 4917–4925. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Chai, X.; Guo, T.; Wu, J.; Yang, P.; Luo, Y.; Zhao, H.; Zhao, W.; Nkechi, O.; Dong, J. Disruption of the Ergosterol Biosynthetic Pathway Results in Increased Membrane Permeability, Causing Overproduction and Secretion of Extracellular Monascus Pigments in Submerged Fermentation. J. Agric. Food Chem. 2019, 67, 13673–13683. [Google Scholar] [CrossRef]
- Hajjaj, H.; Klaébé, A.; Loret, M.O.; Goma, G.; Blanc, P.J.; Fran Ois, J. Biosynthetic Pathway of Citrinin in the Filamentous Fungus Monascus ruber as Revealed by 13C Nuclear Magnetic Resonance. Appl. Environ. Microbiol. 1999, 65, 311–314. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Chen, S.; Gao, M.; Ding, B.; Zhang, J.; Zhou, Y.; Liu, Y.; Yang, H.; Wu, Q.; Chen, F. Acidic conditions induce the accumulation of orange Monascus pigments during liquid-state fermentation of Monascus ruber M7. Appl. Microbiol. Biotechnol. 2019, 103, 8393–8402. [Google Scholar] [CrossRef]
- Liao, Z.; Yang, X.; Fu, H.; Wang, J. The significance of aspartate on NAD(H) biosynthesis and ABE fermentation in Clostridium acetobutylicum ATCC 824. AMB Express 2019, 9, 142. [Google Scholar] [CrossRef]
- Liu, J.; Guo, T.; Wang, D.; Shen, X.; Liu, D.; Niu, H.; Liang, L.; Ying, H. Enhanced butanol production by increasing NADH and ATP levels in Clostridium beijerinckii NCIMB 8052 by insertional inactivation of Cbei_4110. Appl. Microbiol. Biotechnol. 2016, 100, 4985–4996. [Google Scholar] [CrossRef]
- Zhang, X.; Xue, C.; Zhao, F.; Li, D.; Yin, J.; Zhang, C.; Caiyin, Q.; Lu, W. Suitable extracellular oxidoreduction potential inhibit rex regulation and effect central carbon and energy metabolism in Saccharopolyspora spinosa. Microb. Cell Factories 2014, 13, 98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, P.; Huang, J.; Chen, L.; Ding, Z.; Liu, L.; Molnar, I.; Zhang, B. Oxidative stress induction is a rational strategy to enhance the productivity of Antrodia cinnamomea fermentations for the antioxidant secondary metabolite antrodin C. J. Agric. Food Chem. 2020, 68, 3995–4004. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Tan, H.; Lu, F.; Chen, G.; Wu, Z. Changing oxidoreduction potential to improve water-soluble yellow pigment production with Monascus ruber CGMCC 10910. Microb. Cell Factories 2017, 16, 208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nielsen, J. It Is All about Metabolic Fluxes. J. Bacteriol. 2003, 185, 7031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Efremov, R.G.; Baradaran, R.; Sazanov, L.A. The architecture of respiratory complex I. Nature 2010, 465, 441–445. [Google Scholar] [CrossRef]
- Liu, J.; Wu, J.; Cai, X.; Zhang, S.; Lin, Q. Regulation of secondary metabolite biosynthesis in Monascus purpureus via cofactor metabolic engineering strategies. Food Microbiol. 2021, 95, 103689. [Google Scholar] [CrossRef]
- Chai, X.; Ai, Z.; Liu, J.; Guo, T.; Wu, J.; Bai, J.; Lin, Q. Effects of pigment and citrinin biosynthesis on the metabolism and morphology of Monascus purpureus in submerged fermentation. Food Sci. Biotechnol. 2020, 29, 927–937. [Google Scholar] [CrossRef]
- Liu, J.; Guo, T.; Luo, Y.; Chai, X.; Wu, J.; Zhao, W.; Jiao, P.; Luo, F.; Lin, Q. Enhancement of Monascus pigment productivity via a simultaneous fermentation process and separation system using immobilized-cell fermentation. Bioresour. Technol. 2019, 272, 552–560. [Google Scholar] [CrossRef]
- Zhou, B.; Yang, J.; Bi, L.; Li, J.; Ma, Y.; Tian, Y.; Zhong, H.; Ren, J. Quantitative Proteomics Analysis by Sequential Window Acquisition of All Theoretical Mass Spectra–Mass Spectrometry Reveals a Cross-Protection Mechanism for Monascus To Tolerate High-Concentration Ammonium Chloride. J. Agric. Food Chem. 2020, 68, 6672–6682. [Google Scholar] [CrossRef]
- Procaccio, V.; Depetris, D.; Soularue, P.; Mattei, M.G.; Issartel, J.P. cDNA sequence and chromosomal localization of the NDUFS8 human gene coding for the 23 kDa subunit of the mitochondrial complex I. Biochim. Biophys. Acta 1997, 1351, 37–47. [Google Scholar] [CrossRef]
- Sazanov, L.A. Respiratory complex I: Mechanistic and structural insights provided by the crystal structure of the hydrophilic domain. Biochemistry 2007, 46, 2275–2288. [Google Scholar] [CrossRef] [PubMed]
- Baradaran, R.; Berrisford, J.M.; Minhas, G.S.; Sazanov, L.A. Crystal structure of the entire respiratory complex I. Nature 2013, 494, 443–448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, A.; Liu, R.; Miao, Z.-G.; Zhang, X.; Cao, P.-F.; Chen, T.-X.; Li, C.-Y.; Shi, L.; Jiang, A.-L.; Zhao, M.-W. Hydrogen-rich water regulates effects of ROS balance on morphology, growth and secondary metabolism via glutathione peroxidase in Ganoderma lucidum. Environ. Microbiol. 2017, 19, 566–583. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; McMillan-Ward, E.; Kong, J.; Israels, S.J.; Gibson, S.B. Mitochondrial electron-transport-chain inhibitors of complexes I and II induce autophagic cell death mediated by reactive oxygen species. J. Cell Sci. 2007, 120 Pt 23, 4155–4166. [Google Scholar] [CrossRef] [Green Version]
- González-Siso, M.I.; García-Leiro, A.; Tarrío, N.; Cerdán, M.E. Sugar metabolism, redox balance and oxidative stress response in the respiratory yeast Kluyveromyces lactis. Microb. Cell Factories 2009, 8, 46. [Google Scholar] [CrossRef] [Green Version]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef]
- Lu, P.; Wu, A.; Zhang, S.; Bai, J.; Guo, T.; Lin, Q.; Liu, J. Triton X-100 supplementation regulates growth and secondary metabolite biosynthesis during in-depth extractive fermentation of Monascus purpureus. J. Biotechnol. 2021, 341, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Foreman, J.; Demidchik, V.; Bothwell, J.H.; Mylona, P.; Miedema, H.; Torres, M.A.; Linstead, P.; Costa, S.; Brownlee, C.; Jones, J.D.; et al. Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 2003, 422, 442–446. [Google Scholar] [CrossRef]
- Lara-Ortíz, T.; Riveros-Rosas, H.; Aguirre, J. Reactive oxygen species generated by microbial NADPH oxidase NoxA regulate sexual development in Aspergillus nidulans. Mol. Microbiol. 2003, 50, 1241–1255. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Xiong, L.B.; Tao, X.Y.; Liu, Q.H.; Wang, F.Q.; Wei, D.Z. Integrated Transcriptome and Proteome Studies Reveal the Underlying Mechanisms for Sterol Catabolism and Steroid Production in Mycobacterium neoaurum. J. Agric. Food Chem. 2018, 66, 9147–9157. [Google Scholar] [CrossRef]













| Parameters | Escherichia coli K-12 | Clostridium beijerinckii NCIMB 8052 | Yarrowia lipolytica | M. purpureus HQ1 | M. purpureus LQ-6 (in this Study) |
|---|---|---|---|---|---|
| Length of the protein sequence | 180 | 129 | 229 | 225 | 225 |
| Number of predicted TMHs | 0 | 0 | 0 | 0 | 0 |
| Exp number of AAs in TMHs | 0.28102 | 0.00145 | 0.04973 | 0.03387 | 0.03387 |
| Exp number, first 60 AAs | 0.27782 | 0.00013 | 0.01368 | 0.0259 | 0.0259 |
| Total prob of N-in | 0.02778 | 0.02446 | 0.03104 | 0.03812 | 0.03812 |
| Outside | 1~180 | 1~129 | 1~229 | 1~225 | 1~225 |
| Strain | Object | A1 | A2 | X0 | dx | R2 |
|---|---|---|---|---|---|---|
| M. purpureus LQ-6 | T-MPs | 5.78753 | 126.1309 | 142.87989 | 20.60122 | 0.98793 |
| Y-MPs | 0.05763 | 50.90419 | 131.63568 | 28.16209 | 0.98421 | |
| R-MPs | 0.99312 | 36.22905 | 148.89972 | 19.07795 | 0.99059 | |
| O-MPs | 1.95999 | 43.753 | 143.40017 | 18.79606 | 0.98846 | |
| CT | 0.01955 | 1.99955 | 155.31582 | 29.51801 | 0.99583 | |
| Glucose consumption | 83.92365 | 0.12268 | 70.16283 | 14.13061 | 0.99751 | |
| M. purpureus Δ4971 | T-MPs | 4.58268 | 169.02451 | 141.06142 | 22.22952 | 0.99514 |
| Y-MPs | 0.20583 | 70.8763 | 131.97551 | 24.66908 | 0.99326 | |
| R-MPs | 1.28916 | 42.75523 | 146.74975 | 18.9883 | 0.99005 | |
| O-MPs | 1.75071 | 58.17946 | 145.81863 | 20.22318 | 0.99312 | |
| CT | −0.0636 | 3.89083 | 169.87953 | 38.3883 | 0.99891 | |
| Glucose consumption | 79.3281 | 0.07146 | 67.93989 | 7.09183 | 0.99898 |
| GeneID | Description | CT | Log2 Ratio (Δ4971/LQ-6) | |
|---|---|---|---|---|
| LQ-6 | Δ4971 | |||
| Gen-MPDQ_006632 | ndufs8 | 20.24 | 22.23 | −0.410 |
| Gen-MPDQ_006025 | MpigA | 24.58 | 24.23 | 0.695 |
| Gen-MPDQ_006009 | MpigP | 23.60 | 24.23 | 0.125 |
| Gen-MPDQ_006023 | MpigC | 20.30 | 19.58 | 0.587 |
| Gen-MPDQ_006022 | MpigD | 20.94 | 20.98 | 0.192 |
| Gen-MPDQ_006016 | MpigJ | 19.84 | 19.22 | 0.514 |
| Gen-MPDQ_006018 | MpigH | 24.97 | 26.61 | −0.486 |
| Gen-MPDQ_006019 | MpigG | 22.56 | 25.31 | −0.005 |
| Gen-MPDQ_003567 | CtnsS(pksCT) | 25.64 | 24.07 | 1.40 |
| Gen-MPDQ_003569 | ctnsB | 21.53 | 19.42 | 1.67 |
| Gen-MPDQ_003571 | ctnsD | 23.22 | 21.10 | 1.38 |
| Gen-MPDQ_003574 | ctnsC | 23.35 | 21.17 | 1.47 |
| Gen-MPDQ_001039 | Hybrid PKS-NRPS synthet-aseTAS1 | 21.31 | 19.05 | 0.889 |
| Gen-MPDQ_008094 | Hybrid NRPS/PKS enzyme | 24.12 | 22.18 | 0.151 |
| Gen-MPDQ_001369 | NADH deydrogenase | 19.61 | 22.98 | −0.797 |
| Gen-MPDQ_003356 | ETC complex Ⅰ subunit conserved region-domain-containing protein | 20.59 | 23.07 | −0.531 |
| Gen-MPDQ_002668 | NADH:ubiquinone oxidoreductase | 19.89 | 22.83 | −0.844 |
| Gen-MPDQ_004264 | NADH dehydrogenase | 22.45 | 24.63 | −0.490 |
| Gen-MPDQ_000146 | NADH-ubiquinone oxidoreductase | 18.61 | 20.74 | −0.589 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cai, X.; Zhang, S.; Lin, J.; Wang, Y.; Ye, F.; Zhou, B.; Lin, Q.; Liu, J. Role of the Gene ndufs8 Located in Respiratory Complex I from Monascus purpureus in the Cell Growth and Secondary Metabolites Biosynthesis. J. Fungi 2022, 8, 655. https://doi.org/10.3390/jof8070655
Cai X, Zhang S, Lin J, Wang Y, Ye F, Zhou B, Lin Q, Liu J. Role of the Gene ndufs8 Located in Respiratory Complex I from Monascus purpureus in the Cell Growth and Secondary Metabolites Biosynthesis. Journal of Fungi. 2022; 8(7):655. https://doi.org/10.3390/jof8070655
Chicago/Turabian StyleCai, Xinru, Song Zhang, Jia Lin, Yaxu Wang, Fanyu Ye, Bo Zhou, Qinlu Lin, and Jun Liu. 2022. "Role of the Gene ndufs8 Located in Respiratory Complex I from Monascus purpureus in the Cell Growth and Secondary Metabolites Biosynthesis" Journal of Fungi 8, no. 7: 655. https://doi.org/10.3390/jof8070655