Computational and Preclinical Analysis of 2-(4-Methyl)benzylidene-4,7-dimethyl Indan-1-one (IPX-18): A Novel Arylidene Indanone Small Molecule with Anti-Inflammatory Activity via NF-κB and Nrf2 Signaling
Abstract
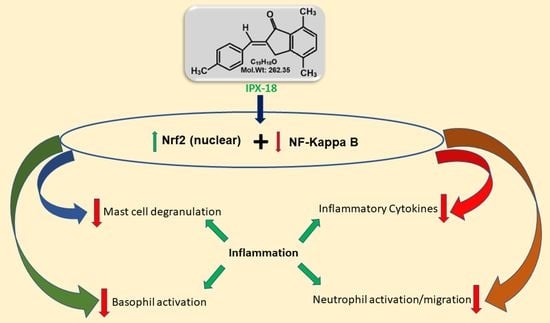
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
2.2.1. Synthesis of 2-(4-Methyl)benzylidene-4,7-dimethyl Indan-1-one
2.2.2. Ethical Approval
2.2.3. Cell Culture
2.2.4. Annexin V Assay for Apoptosis/Cell Death
2.2.5. Cytokine Assays
2.2.6. Isolation of Neutrophils from HWB
2.2.7. Neutrophil Migration Inhibition Assay
2.2.8. Neutrophil Elastase Assay
2.2.9. Basophil Activation Assay (BAT assay) in HWB
2.2.10. Cell Viability Assay
2.2.11. TNF-α and Degranulation in RBL-2H3 Cells
2.2.12. Structure Retrieval and Protein–Ligand Docking
2.2.13. Molecular Dynamic Simulation
2.2.14. Flow Cytometry
2.2.15. Statistical Analysis
3. Results
3.1. Chemistry of Synthesized Small Molecule
3.2. Nontoxicity of IPX-18
3.3. IPX-18 Attenuated Proinflammatory Cytokine Responses in Human Whole Blood and Peripheral Nucleocytes
3.4. Effect of IPX-18 Treatment on Activated Neutrophils
3.5. IPX-18 Effectively Inhibited the Activation of Basophils
3.6. Dose Tolerance of IPX-18 on Normal and Stimulated RBL-2H3 Cells
3.7. IPX-18 Inhibited TNF-α Release and Degranulation in Anti-DNP/IgE of Sensitized RBL-2H3 Cells
3.8. Protein–Ligand Docking of IPX-18 to p50 Subunit of NFkB
3.9. Molecular Dynamic Simulation Predicted IPX-18 Binding Stability
3.10. Efficacy of IPX-18 on Key Signaling Proteins of the Inflammatory Pathway
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Scrivo, R.; Vasile, M.; Bartosiewicz, I.; Valesini, G. Inflammation as “common soil” of the multifactorial diseases. Autoimmun. Rev. 2011, 10, 369–374. [Google Scholar] [CrossRef] [PubMed]
- Tsai, Y.-F.; Yu, H.-P.; Chang, W.-Y.; Liu, F.-C.; Huang, Z.-C.; Hwang, T.-L. Sirtinol Inhibits Neutrophil Elastase Activity and Attenuates Lipopolysaccharide-Mediated Acute Lung Injury in Mice. Sci. Rep. 2015, 5, 8347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cornette, L. Fetal and neonatal inflammatory response and adverse outcome. Semin. Fetal Neonatal Med. 2004, 9, 459–470. [Google Scholar] [CrossRef] [PubMed]
- Doyle, L.W.; Cheong, J.L.; Ehrenkranz, R.A.; Halliday, H.L. Early (<8 days) systemic postnatal corticosteroids for prevention of bronchopulmonary dysplasia in preterm infants. Cochrane Database Syst. Rev. 2017, 2017, CD001146. [Google Scholar]
- Hinz, M.; Scheidereit, C. The IκB kinase complex in NF-κB regulation and beyond. EMBO Rep. 2014, 15, 46–61. [Google Scholar] [CrossRef] [Green Version]
- Saha, S.; Buttari, B.; Panieri, E.; Profumo, E.; Saso, L. An Overview of Nrf2 Signaling Pathway and Its Role in Inflammation. Molecules 2020, 25, 5474. [Google Scholar] [CrossRef]
- Soares, M.P.; Seldon, M.P.; Gregoire, I.P.; Vassilevskaia, T.; Berberat, P.O.; Yu, J.; Tsui, T.-Y.; Bach, F.H. Heme Oxygenase-1 Modulates the Expression of Adhesion Molecules Associated with Endothelial Cell Activation. J. Immunol. 2004, 172, 3553–3563. [Google Scholar] [CrossRef] [Green Version]
- Yerra, V.G.; Negi, G.; Sharma, S.S.; Kumar, A. Potential therapeutic effects of the simultaneous targeting of the Nrf2 and NF-κB pathways in diabetic neuropathy. Redox Biol. 2013, 1, 394–397. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.-G.; Zhang, Y.-Q.; Wu, Z.-Z.; Hsieh, C.-W.; Chu, C.-S.; Wung, B.-S. Peanut arachidin-1 enhances Nrf2-mediated protective mechanisms against TNF-α-induced ICAM-1 expression and NF-κB activation in endothelial cells. Int. J. Mol. Med. 2018, 41, 541–547. [Google Scholar] [CrossRef] [Green Version]
- Liu, G.-H.; Qu, J.; Shen, X. NF-κB/p65 antagonizes Nrf2-ARE pathway by depriving CBP from Nrf2 and facilitating recruitment of HDAC3 to MafK. Biochim. Biophys. Acta 2008, 1783, 713–727. [Google Scholar] [CrossRef] [Green Version]
- Brigelius-Flohé, R.; Flohé, L. Basic principles and emerging concepts in the redox control of transcription factors. Antioxid. Redox Signal. 2011, 15, 2335–2381. [Google Scholar] [CrossRef] [Green Version]
- Bellezza, I.; Mierla, A.L.; Minelli, A. Nrf2 and NF-κB and their concerted modulation in cancer pathogenesis and progression. Cancers 2010, 2, 483–497. [Google Scholar] [CrossRef]
- Beurel, E.; Michalek, S.M.; Jope, R.S. Innate and adaptive immune responses regulated by glycogen synthase kinase-3 (GSK3). Trends Immunol. 2010, 31, 24–31. [Google Scholar] [CrossRef] [Green Version]
- Karrouchi, K.; Radi, S.; Ramli, Y.; Taoufik, J.; Mabkhot, Y.N.; Al-Aizari, F.A.; Ansar, M. Synthesis and Pharmacological Activities of Pyrazole Derivatives: A Review. Molecules 2018, 23, 134. [Google Scholar] [CrossRef] [Green Version]
- Abdelrazek, F.M.; Metz, P.; Kataeva, O.; Jäger, A.; El-Mahrouky, S.F. Synthesis and molluscicidal activity of new chromene and pyrano [2,3-c]pyrazole derivatives. Arch. Pharm. 2007, 340, 543–548. [Google Scholar] [CrossRef]
- Ottanà, R.; Maccari, R.; Ciurleo, R.; Vigorita, M.G.; Panico, A.M.; Cardile, V.; Garufi, F.; Ronsisvalle, S. Synthesis and in vitro evaluation of 5-arylidene-3-hydroxyalkyl-2-phenylimino-4-thiazolidinones with antidegenerative activity on human chondrocyte cultures. Bioorg. Med. Chem. 2007, 15, 7618–7625. [Google Scholar] [CrossRef]
- da Costa Leite, L.F.; Veras Mourão, R.H.; Alves de Lima, M.d.C.; Galdino, S.L.; Hernandes, M.Z.; Rocha Neves, F.d.A.; Vidal, S.; Barbe, J.; da Rocha Pitta, I. Synthesis, biological evaluation and molecular modeling studies of arylidene-thiazolidinediones with potential hypoglycemic and hypolipidemic activities. Eur. J. Med. Chem. 2007, 42, 1263–1271. [Google Scholar] [CrossRef]
- Xiao, S.; Zhang, W.; Chen, H.; Fang, B.; Qiu, Y.; Chen, X.; Chen, L.; Shu, S.; Zhang, Y.; Zhao, Y.; et al. Design, synthesis, and structure–activity relationships of 2-benzylidene-1-indanone derivatives as anti-inflammatory agents for treatment of acute lung injury. Drug Des. Dev. Ther. 2018, 12, 887–899. [Google Scholar] [CrossRef] [Green Version]
- Juthani, V.V.; Clearfield, E.; Chuck, R.S. Non-steroidal anti-inflammatory drugs versus corticosteroids for controlling inflammation after uncomplicated cataract surgery. Cochrane Database Syst. Rev. 2017, 2017, CD010516. [Google Scholar] [CrossRef]
- Corkum, C.P.; Ings, D.P.; Burgess, C.; Karwowska, S.; Kroll, W.; Michalak, T.I. Immune cell subsets and their gene expression profiles from human PBMC isolated by Vacutainer Cell Preparation Tube (CPT™) and standard density gradient. BMC Immunol. 2015, 16, 48. [Google Scholar] [CrossRef] [Green Version]
- Maqbool, M.; Vidyadaran, S.; George, E.; Ramasamy, R. Optimisation of laboratory procedures for isolating human peripheral blood derived neutrophils. Med. J. Malays. 2011, 66, 296–299. [Google Scholar]
- Craciun, I.; Fenner, A.M.; Kerns, R.J. N-Arylacyl O-sulfonated aminoglycosides as novel inhibitors of human neutrophil elastase, cathepsin G and proteinase 3. Glycobiology 2016, 26, 701–709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dera, A.; Rajagopalan, P. Thymoquinone attenuates phosphorylation of AKT to inhibit kidney cancer cell proliferation. J. Food Biochem. 2019, 43, e12793. [Google Scholar] [CrossRef] [PubMed]
- Naal, R.M.Z.; Tabb, J.; Holowka, D.; Baird, B. In situ measurement of degranulation as a biosensor based on RBL-2H3 mast cells. Biosens. Bioelectron. 2004, 20, 791–796. [Google Scholar] [CrossRef]
- Al Shahrani, M.; Abohassan, M.; Alshahrani, M.Y.; Hakami, A.R.; Rajagopalan, P. High-throughput virtual screening and preclinical analysis identifies CB-1, a novel potent dual B-Raf/c-Raf inhibitor, effective against wild and mutant variants of B-Raf expression in colorectal carcinoma. J. Comput.-Aided Mol. Des. 2021, 35, 1165–1176. [Google Scholar] [CrossRef]
- Kamli, H.; Zaman, G.S.; Shaikh, A.; Mobarki, A.A.; Rajagopalan, P. A Combined Chemical, Computational, and In Vitro Approach Identifies SBL-105 as Novel DHODH Inhibitor in Acute Myeloid Leukemia Cells. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2022, 28, 899–911. [Google Scholar] [CrossRef]
- Ahmad, I.; Irfan, S.; Ali Beg, M.M.; Kamli, H.; Ali, S.P.; Begum, N.; Alshahrani, M.Y.; Rajagopalan, P. The SMAC mimetic AT-101 exhibits anti-tumor and anti-metastasis activity in lung adenocarcinoma cells by the IAPs/caspase-dependent apoptosis and p65-NFƙB cross-talk. Iran. J. Basic Med. Sci. 2021, 24, 969–977. [Google Scholar]
- Rajakariar, R.; Yaqoob, M.M.; Gilroy, D.W. COX-2 in inflammation and resolution. Mol. Interv. 2006, 6, 199–207. [Google Scholar] [CrossRef]
- Bozinovski, S.; Jones, J.E.; Vlahos, R.; Hamilton, J.A.; Anderson, G.P. Granulocyte/macrophage-colony-stimulating factor (GM-CSF) regulates lung innate immunity to lipopolysaccharide through Akt/Erk activation of NFκB and AP-1 in vivo. J. Biol. Chem. 2002, 277, 42808–42814. [Google Scholar] [CrossRef] [Green Version]
- Stafford, J.L.; Neumann, N.F.; Belosevic, M. Macrophage-Mediated Innate Host Defense Against Protozoan Parasites. Crit. Rev. Microbiol. 2002, 28, 187–248. [Google Scholar] [CrossRef]
- Kayhan, S.; Guzel, A.; Duran, L.; Tutuncu, S.; Guzel, A.; Gunaydın, M.; Salis, O.; Okuyucu, A.; Selcuk, M.Y. Effects of leflunomide on inflamation and fibrosis in bleomycine induced pulmonary fibrosis in wistar albino rats. J. Thorac. Dis. 2013, 5, 641–649. [Google Scholar] [CrossRef]
- Sedgwick, J.B.; Menon, I.; Gern, J.E.; Busse, W.W. Effects of inflammatory cytokines on the permeability of human lung microvascular endothelial cell monolayers and differential eosinophil transmigration. J. Allergy Clin. Immunol. 2002, 110, 752–756. [Google Scholar] [CrossRef]
- Gabay, C. Interleukin-6 and chronic inflammation. Arthritis Res. Ther. 2006, 8, S3. [Google Scholar] [CrossRef] [Green Version]
- Doherty, D.E.; Downey, G.P.; Worthen, G.S.; Haslett, C.; Henson, P.M. Monocyte retention and migration in pulmonary inflammation. Requirement for neutrophils. Lab. Investig. J. Tech. Methods Pathol. 1988, 59, 200–213. [Google Scholar]
- Oliveira, S.H.; Canetti, C.; Ribeiro, R.A.; Cunha, F.Q. Neutrophil Migration Induced by IL-1β Depends upon LTB4 Released by Macrophages and upon TNF-α and IL-1β Released by Mast Cells. Inflammation 2008, 31, 36–46. [Google Scholar] [CrossRef]
- Driessler, F.; Venstrom, K.; Sabat, R.; Asadullah, K.; Schottelius, A.J. Molecular mechanisms of interleukin-10-mediated inhibition of NF-κB activity: A role for p50. Clin. Exp. Immunol. 2004, 135, 64–73. [Google Scholar] [CrossRef]
- Siracusa, M.C.; Kim, B.S.; Spergel, J.M.; Artis, D. Basophils and allergic inflammation. J. Allergy Clin. Immunol. 2013, 132, 789–801. [Google Scholar] [CrossRef] [Green Version]
- Knol, E.F.; Mul, F.P.; Jansen, H.; Calafat, J.; Roos, D. Monitoring human basophil activation via CD63 monoclonal antibody 435. J. Allergy Clin. Immunol. 1991, 88, 328–338. [Google Scholar] [CrossRef]
- Wedi, B.; Kapp, A. Cellular in-vitro assays. Applicability in daily routine. Der Hautarzt Z. Fur Dermatol. Vener-Ologie Und Verwandte Geb. 2010, 61, 954–960. [Google Scholar] [CrossRef]
- Hausmann, O.V.; Gentinetta, T.; Fux, M.; Ducrest, S.; Pichler, W.J.; Dahinden, C.A. Robust expression of CCR3 as a single basophil selection marker in flow cytometry. Allergy 2011, 66, 85–91. [Google Scholar] [CrossRef]
- Sturm, G.J.; Kranzelbinder, B.; Sturm, E.M.; Heinemann, A.; Groselj-Strele, A.; Aberer, W. The basophil activation test in the diagnosis of allergy: Technical issues and critical factors. Allergy 2009, 64, 1319–1326. [Google Scholar] [CrossRef] [PubMed]
- Özdemir, S.K.; Güloğlu, D.; Sin, B.A.; Elhan, A.H.; İkincioğulları, A.; Mısırlıgil, Z. Reliability of basophil activation test using CD203c expression in diagnosis of pollen allergy. Am. J. Rhinol. Allergy 2011, 25, e225–e231. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Zhang, H.; Zeng, X.; Yang, P. Self-amplification mechanisms of mast cell activation: A new look in allergy. Curr. Mol. Med. 2012, 12, 1329–1339. [Google Scholar] [CrossRef] [PubMed]
- Kolaczkowska, E.; Kubes, P. Neutrophil recruitment and function in health and inflammation. Nat. Rev. Immunol. 2013, 13, 159–175. [Google Scholar] [CrossRef] [PubMed]
- Park, E.K.; Jung, H.S.; Yang, H.I.; Yoo, M.C.; Kim, C.; Kim, K.S. Optimized THP-1 differentiation is required for the detection of responses to weak stimuli. Inflamm. Res. Off. J. Eur. Histamine Res. Soc. 2007, 56, 45–50. [Google Scholar] [CrossRef]
- Giridharan, S.; Srinivasan, M. Mechanisms of NF-κB p65 and strategies for therapeutic manipulation. J. Inflamm. Res. 2018, 11, 407–419. [Google Scholar] [CrossRef] [Green Version]
- Yan, T.; Yu, X.; Sun, X.; Meng, D.; Jia, J.-M. A new steroidal saponin, furotrilliumoside from Trillium tschonoskii inhibits lipopolysaccharide-induced inflammation in Raw264.7 cells by targeting PI3K/Akt, MARK and Nrf2/HO-1 pathways. Fitoterapia 2016, 115, 37–45. [Google Scholar] [CrossRef]








Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gahtani, R.M.; Shaikh, A.; Kamli, H. Computational and Preclinical Analysis of 2-(4-Methyl)benzylidene-4,7-dimethyl Indan-1-one (IPX-18): A Novel Arylidene Indanone Small Molecule with Anti-Inflammatory Activity via NF-κB and Nrf2 Signaling. Biomedicines 2023, 11, 716. https://doi.org/10.3390/biomedicines11030716
Gahtani RM, Shaikh A, Kamli H. Computational and Preclinical Analysis of 2-(4-Methyl)benzylidene-4,7-dimethyl Indan-1-one (IPX-18): A Novel Arylidene Indanone Small Molecule with Anti-Inflammatory Activity via NF-κB and Nrf2 Signaling. Biomedicines. 2023; 11(3):716. https://doi.org/10.3390/biomedicines11030716
Chicago/Turabian StyleGahtani, Reem M., Ahmad Shaikh, and Hossam Kamli. 2023. "Computational and Preclinical Analysis of 2-(4-Methyl)benzylidene-4,7-dimethyl Indan-1-one (IPX-18): A Novel Arylidene Indanone Small Molecule with Anti-Inflammatory Activity via NF-κB and Nrf2 Signaling" Biomedicines 11, no. 3: 716. https://doi.org/10.3390/biomedicines11030716