Structural Basis of the Interaction of the G Proteins, Gαi1, Gβ1γ2 and Gαi1β1γ2, with Membrane Microdomains and Their Relationship to Cell Localization and Activity
Abstract
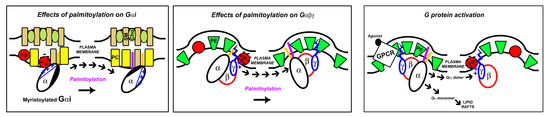
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Site-Directed Mutagenesis and Cloning of G Proteins
2.3. G Protein Purification
2.3.1. Gαi1 Proteins
2.3.2. Gβ1γ2 Dimers
2.3.3. Gαi1β1γ2 Heterotrimers
2.4. Acylation Reaction in Gαi1β1γ2 Heterotrimers
2.5. G Protein Binding to Model Membranes
2.6. G Protein Structure Analysis
2.7. Data Analysis
3. Results
3.1. Membranes Used in the Present Study
3.2. The Role of Gγ2 C-Terminal Region in Gβ1γ2–Membrane Interactions
3.2.1. Geranylgeranyl Is Critical for the Membrane Binding of Gβ1γ2
3.2.2. Geranylgeranyl plus the Neighboring Basic Amino Acids Arg-62 and Lys-65 Drive Gβ1γ2 towards PE-Rich (Non-Lamellar Prone) Ld Membrane Microdomains
3.2.3. Gγ2 C-Terminal Basic Amino Acids Drive the Interaction of Gβ1γ2 with PS-Rich Membranes
3.2.4. Geranylgeranyl plus the Basic Arg-62, Lys-64 and Lys-65 Amino Acids Strongly Modulate the Interaction of Gβ1γ2 with PE- and PS-Rich Membranes
3.2.5. Arg-62 and Lys-65 Are Critical Residues in the Interaction between Gβ1γ2 and Ordered Lamellar Membranes
3.2.6. Geranylgeranylation Drives the Localization of Gγ2 and Gβ1γ2 to Biological Membranes
3.3. Effects of the Gγ2 C-Terminal and Gαi1 N-Terminal Regions and Membrane Lipid Organization on Gαi1β1γ2-Membrane Interactions
3.3.1. Geranylgeranyl and Myristoyl Moieties Are Required for Gαi1β1γ2 Targeting to PE-Rich Non-Lamellar Prone Microdomains
3.3.2. Gαi1 Myristoylation and Palmitoylation and the Gγ2 C-Terminal Polybasic Domain Regulate Gαi1β1γ2-PS Interactions
3.3.3. Myristoylated and Non-Palmitoylated Gαi1β1γ2 Complexes Have a High Affinity for PE- and PS-Rich Membrane Microdomains
3.3.4. Gαi1 Myristic Acid and C-Terminal Gγ2 Basic Amino Acids Prevent Gαi1β1γ2 Targeting to Raft-like Membrane Domains
3.4. The Gαi1 Monomer and the Corresponding Heterotrimer Differ Remarkably in Their Binding to Membranes
3.5. The Myristoyl and Geranylgeranyl Moieties Plus the Gγ2 C-Terminal Polybasic Domain Are Key Determinants of the Interaction of Gαi1β1γ2 with Biological Membranes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Escribá, P.V.; Wedegaertner, P.B.; Goñi, F.M.; Vögler, O. Lipid-protein interactions in GPCR-associated signaling. Biochim. Biophys. Acta 2007, 1768, 836–852. [Google Scholar] [CrossRef]
- Vögler, O.; Barceló, J.M.; Ribas, C.; Escribá, P.V. Membrane interactions of G proteins and other related proteins. Biochim. Biophys. Acta 2008, 1778, 1640–1652. [Google Scholar] [CrossRef]
- Dupré, D.J.; Robitaille, M.; Rebois, R.V.; Hébert, T.E. The Role of Gβγ Subunits in the Organization, Assembly, and Function of GPCR Signaling Complexes. Annu. Rev. Pharmacol. Toxicol. 2009, 49, 31–56. [Google Scholar] [CrossRef] [PubMed]
- Syrovatkina, V.; Alegre, K.O.; Dey, R.; Huang, X.Y. Regulation, signaling, and physiological functions of G-Proteins. J. Mol. Biol. 2016, 428, 3850–3868. [Google Scholar] [CrossRef]
- López, D.J.; Alvarez, R.; Escribá, P.V. Lipid-protein interactions in G protein signal transduction. In G Protein-Coupled Receptors: From Structure to Function, 1st ed.; RSC Publishing: Cambridge, UK, 2011; pp. 153–178. [Google Scholar]
- Noguera-Salvà, M.A.; Guardiola-Serrano, F.; Martin, M.L.; Marcilla-Etxenike, A.; Bergo, M.O.; Busquets, X.; Escribá, P.V. Role of the C-terminal basic amino acids and the lipid anchor of the Gγ2 protein in membrane interactions and cell localization. Biochim. Biophys. Acta 2017, 1859, 1536–1547. [Google Scholar] [CrossRef]
- Jones, T.L.; Simonds, W.F.; Merendino, J.J., Jr.; Brann, M.R.; Spiegel, A.M. Myristoylation of an inhibitory GTP-binding protein alpha subunit is essential for its membrane attachment. Proc. Natl. Acad. Sci. USA 1990, 87, 568–572. [Google Scholar] [CrossRef]
- Mumby, S.M.; Casey, P.J.; Gilman, A.G.; Gutowski, S.; Sternweis, P.C. G protein gamma subunits contain a 20-carbon isoprenoid. Proc. Natl. Acad. Sci. USA 1990, 87, 5873–5877. [Google Scholar] [CrossRef]
- Linder, M.E.; Middleton, P.; Hepler, J.R.; Taussig, R.; Gilman, A.G.; Mumby, S.M. Lipid modifications of G proteins: α subunits are palmitoylated. Proc. Natl. Acad. Sci. USA 1993, 90, 3675–3679. [Google Scholar] [CrossRef] [PubMed]
- Alves, I.D.; Lecomte, S. Study of G-Protein Coupled Receptor Signaling in Membrane Environment by Plasmon Waveguide Resonance. Acc. Chem. Res. 2019, 52, 1059–1067. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Luttrell, L.M.; Premont, R.T.; Rockey, D.C. β-Arrestin is a critical component of the GPCR-eNos signalosome. Proc. Natl. Acad. Sci. USA 2020, 117, 11483–11492. [Google Scholar] [CrossRef]
- Hanyaloglu, A.C. Advances in membrane trafficking and endosomal signaling of G protein-coupled receptors. Int. Rev. Cell Mol. Biol. 2018, 339, 93–131. [Google Scholar]
- Zhang, H.; Ping, H.; Zhou, Q.; Lu, Y.; Lu, B. The potential oncogenic and MLN4924-resistant effects of CSN5 on cervical cancer cells. Cancer Cell Int. 2021, 21, 369. [Google Scholar] [CrossRef]
- Singer, S.J.; Nicolson, G. The fluid mosaic model of the structure of cell membranes. Science 1972, 175, 720–731. [Google Scholar] [CrossRef] [PubMed]
- Vereb, G.; Szollosi, J.; Matko, J.; Nagy, P.; Farkas, T.; Vigh, L.; Matyus, L.; Waldmann, T.A.; Damjanovich, S. Dynamic, yet structured: The cell membrane three decades after the Singer-Nicolson model. Proc. Natl. Acad. Sci. USA 2003, 100, 8053–8058. [Google Scholar] [CrossRef] [PubMed]
- Escribá, P.V. Membrane-lipid therapy: A new approach in molecular medicine. Trends Mol. Med. 2006, 12, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Escribá, P.V.; González-Ros, J.M.; Goñi, F.M.; Kinnunen, P.K.; Vigh, L.; Sánchez-Magraner, L.; Fernández, A.M.; Busquets, X.; Horváth, I. Membranes: A meeting point for lipids, proteins and therapies. J. Cell. Mol. Med. 2008, 12, 829–875. [Google Scholar] [CrossRef]
- Lingwood, D.; Simons, K. Lipid rafts as a membrane-organizing principle. Science 2010, 327, 46–50. [Google Scholar] [CrossRef]
- Nicolson, G.L. The fluid-mosaic model of membrane structure: Still relevant to understanding the structure, function and dynamics of biological membranes after more than 40 years. Biochim. Biophys. Acta 2014, 1838, 1451–1466. [Google Scholar] [CrossRef]
- Torres, M.; Rosselló, C.A.; Fernáncez-García, P.; Lladó, V.; Kakhlon, O.; Escribá, P.V. The implications for cells of the lipid switches driven by protein-membrane interactions and the development of membrane lipid therapy. Int. J. Mol. Sci. 2019, 21, 2322. [Google Scholar] [CrossRef]
- Israelachvili, J.N.; Marcelja, S.; Horn, R.G. Physical principles of membrane organisation. Quart. Rev. Biophys. 1980, 13, 121–200. [Google Scholar] [CrossRef]
- Escribá, P.V.; Sastre, M.; García-Sevilla, J.A. Disruption of cellular signaling pathways by daunomycin through destabilization of nonlamellar membrane structures. Proc. Natl. Acad. Sci. USA 1995, 92, 7595–7599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Escribá, P.V.; Ozaita, A.; Ribas, C.; Miralles, A.; Fodor, E.; Farkas, T.; García-Sevilla, J.A. Role of lipid polymorphism in G protein-membrane interactions: Nonlamellar-prone phospholipids and peripheral protein binding to membranes. Proc. Natl. Acad. Sci. USA 1997, 94, 11375–11380. [Google Scholar] [CrossRef] [PubMed]
- Vögler, O.; Casas, J.; Capó, D.; Nagy, T.; Borchert, G.; Martorell, G.; Escribá, P.V. The Gβγ dimer drives the interaction of heterotrimeric Gi proteins with nonlamellar membrane structures. J. Biol. Chem. 2004, 279, 36540–36545. [Google Scholar] [CrossRef]
- Melkonian, K.A.; Ostermeyer, A.G.; Chen, J.Z.; Roth, M.G.; Brown, D.A. Role of lipid modifications in targeting proteins to detergent-resistant membrane rafts. Many raft proteins are acylated, while few are prenylated. J. Biol. Chem. 1999, 274, 3910–3917. [Google Scholar] [CrossRef]
- Lu, X.; Sicard, R.; Jiang, X.; Stockus, J.N.; McNamara, G.; Abdulreda, M.; Moy, V.T.; Landgraf, R.; Lossos, I.S. HGAL localization to cell membrane regulates B-cell receptor signaling. Blood 2015, 125, 649–657. [Google Scholar] [CrossRef] [PubMed]
- Moffett, S.; Brown, D.A.; Linder, M.E. Lipid-dependent targeting of G proteins into rafts. J. Biol. Chem. 2000, 275, 2191–2198. [Google Scholar] [CrossRef]
- Zhou, Y.; Prakash, P.S.; Liang, H.; Gorfe, A.A.; Hancock, J.F. The KRAS and other prenylated polybasic domain membrane anchors recognize phosphatydilserine acyl chain structure. Proc. Natl. Acad. Sci. USA 2021, 118, e2014605118. [Google Scholar] [CrossRef]
- Koklič, T.; Hrovat, A.; Guixà-González, R.; Rodríguez-Espigares, I.; Navio, D.; Frangež, R.; Uršič, M.; Kubale, V.; Plemenitaš, A.; Selent, J.; et al. Electron paramagnetic resonance gives evidence for the presence of Type 1 Gonadotropin-Releasing Hormone Receptor (GnRH-R) in subdomains of lipid rafts. Molecules 2021, 26, 973. [Google Scholar] [CrossRef]
- Eggeling, C.; Ringemann, C.; Medda, R.; Schwarzmann, G.; Sandhoff, K.; Polyakova, S.; Belov, V.N.; Hein, B.; von Middendorff, C.; Schönle, A.; et al. Direct observation of the nanoscale dynamic of membrane lipids in a living cell. Nature 2009, 457, 1159–1162. [Google Scholar] [CrossRef]
- Sot, J.; Ibarguren, M.; Busto, J.V.; Montes, L.R.; Goñi, F.M.; Alonso, A. Cholesterol displacement by ceramide in sphingomyelin-containing liquid-ordered domains, and generation of gel regions in giant lipidic vesicles. FEBS Lett. 2008, 582, 3230–3236. [Google Scholar] [CrossRef]
- Rothman, J.E.; Lenard, J. Membrane asymmetry. Science 1977, 195, 743–753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hessel, E.; Heck, M.; Müller, P.; Herrmann, A.; Hofmann, K.P. Signal Transduction in the Visual Cascade Involves Specific Lipid-Protein Interactions. J. Biol. Chem. 2003, 278, 22853–22860. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Wong, C.; Cho, K.; van der Hoeven, D.; Liang, H.; Thakur, D.P.; Luo, J.; Babic, M.; Zinsmaier, K.E.; Zhu, M.X.; et al. Membrane potential modulates plasma membrane phospholipid dynamics and K-Ras signaling. Science 2015, 349, 873–876. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, R.; López, D.J.; Casas, J.; Lladó, V.; Higuera, M.; Nagy, T.; Barceló, M.; Busquets, X.; Escribá, P.V. G protein-membrane interactions I: Gαi1 myristoyl and palmitoyl modifications in protein-lipid protein–lipid interactions and its implications in membrane microdomain localization. Biochim. Biophys. Acta 2015, 1851, 1511–1520. [Google Scholar] [CrossRef]
- Casas, J.; Ibarguren, M.; Álvarez, R.; Terés, S.; Lladó, V.; Piotto, S.; Concilio, S.; Busquets, X.; López, D.J.; Escribá, P.V. G protein-membrane interactions II: Effect of G protein-linked lipids on membrane structure and G protein-membrane interactions. Biochim. Biophys. Acta 2017, 1859, 1526–1535. [Google Scholar] [CrossRef]
- Damian, M.; Louet, M.; Gomes, A.A.S.; M’Kadmi, C.; Denoyelle, S.; Cantel, S.; Mary, S.; Bisch, P.M.; Fehrentz, J.-A.; Catoire, L.J.; et al. Allosteric modulation of ghrelin receptor signaling by lipids. Nat. Commun. 2021, 12, 3938. [Google Scholar] [CrossRef]
- Escribá, P.V. Membrane-lipid therapy: A historical perspective of membrane-targeted therapies—From lipid bilayer structure to the pathophysiological regulation of cells. Biochim. Biophys. Acta 2017, 1859, 1493–1506. [Google Scholar] [CrossRef]
- Maldonado, C.; Nguyen, M.-D.; Bauer, P.; Nakamura, S.; Khundmiri, S.; Perez-Abadia, G.; Stowers, H.L.; Wu, W.-J.; Tang, X.-L. Rapid lipid modification of endothelial cell membranes in cardiac ischemia/reperfusion injury: A novel therapeutic strategy to reduce infarct size. Cardiovasc. Drugs Ther. 2021, 35, 113–123. [Google Scholar] [CrossRef]
- Kozasa, T.; Gilman, A.G. Purification of recombinant G proteins from Sf9 cells by hexahistidine tagging of associated subunits. J. Biol. Chem. 1995, 270, 1734–1741. [Google Scholar] [CrossRef]
- García-Sevilla, J.A.; Escribá, P.V.; Ozaita, A.; La Harpe, R.; Walzer, C.; Eytan, A.; Guimón, J. Regulation of immunolabeled α2A-adrenoceptors, Gi coupling proteins, and regulatory receptor kinases in the prefrontal cortex of depressed suicides. J. Neurochem. 1999, 72, 282–291. [Google Scholar] [CrossRef]
- Escribá, P.V.; Sánchez-Domínguez, J.M.; Alemany, R.; Perona, J.S.; Ruiz-Gutierrez, V. Alteration of lipids, G proteins, and PKC in cell membranes of elderly hypertensives. Hypertension 2003, 41, 176–182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barceló, F.; Prades, J.; Encinar, J.A.; Funari, S.S.; Vögler, O.; González-Ros, J.M.; Escribá, P.V. Interaction of the C-terminal region of the Gg protein with model membranes. Biophys. J. 2007, 93, 2530–2541. [Google Scholar] [CrossRef]
- Terés, S.; Barceló-Coblijn, G.; Benet, M.; Alvarez, R.; Bressani, R.; Halver, J.E.; Escribá, P.V. Oleic acid content is responsible for the reduction in blood pressure induced by olive oil. Proc. Natl. Acad. Sci. USA 2008, 105, 13811–13816. [Google Scholar] [CrossRef] [PubMed]
- Ibarguren, M.; López, D.J.; Encinar, J.A.; González-Ros, J.M.; Busquets, X.; Escribá, P.V. Partitioning of liquid-ordered/liquid-disordered membrane microdomains induced by the fluidifying effect of 2-hydroxylated fatty acid derivatives. Biochim. Biophys. Acta 2013, 1828, 2553–2563. [Google Scholar] [CrossRef] [PubMed]
- Ibarguren, M.; López, D.J.; Escribá, P.V. The effect of natural and synthetic fatty acids on membrane structure, microdomain organization, cellular functions and human health. Biochim. Biophys. Acta 2014, 1838, 1518–1528. [Google Scholar] [CrossRef]
- Terés, S.; Lladó, V.; Higuera, M.; Barceló-Coblijn, G.; Martin, M.L.; Noguera-Salvà, M.A.; Marcilla-Etxenike, A.; García-Verdugo, J.M.; Soriano-Navarro, M.; Saus, C.; et al. 2-Hydroxyoleate, a nontoxic membrane binding anticancer drug, induces glioma cell differentiation and autophagy. Proc. Natl. Acad. Sci. USA 2012, 109, 8489–8494. [Google Scholar] [CrossRef] [PubMed]
- Escribá, P.V.; Busquets, X.; Inokuchi, J.-I.; Balogh, G.; Török, Z.; Horváth, I.; Harwood, J.L.; Vígh, L. Membrane lipid therapy: Modulation of the cell membrane composition and structure as a molecular base for drug discovery and new disease treatment. Prog. Lipid Res. 2015, 59, 38–53. [Google Scholar] [CrossRef]
- Wall, M.A.; Coleman, D.E.; Lee, E.; Iñiguez-Lluhi, J.A.; Posner, B.A.; Gilman, A.G.; Sprang, S.R. The Structure of the G Protein Heterotrimer Giα1β1γ2. Cell 1995, 83, 1047–1058. [Google Scholar] [CrossRef]
- Lambright, D.G.; Sondek, J.; Bohm, A.; Skiba, N.P.; Hamm, H.E.; Sigler, P.B. The 2.0 A crystal structure of a heterotrimeric G protein. Nature 1996, 379, 311–319. [Google Scholar] [CrossRef]
- van Keulen, S.C.; Rothlisberger, U. Exploring the inhibition mechanism of adenylyl cyclase type 5 by n-terminal myristoylated Gαi1. PLoS Comput. Biol. 2017, 13, e1005673. [Google Scholar] [CrossRef]
- Degtyarev, M.Y.; Spiegel, A.M.; Jones, T.L.Z. Palmitoylation of a G Protein αi Subunit Requires Membrane Localization Not Myristoylation. J. Biol. Chem. 1994, 269, 30898–30903. [Google Scholar] [CrossRef]
- Bernstein, L.S.; Grillo, A.A.; Loranger, S.S.; Linder, M.E. RGS4 binds to membranes through an amphipathic alpha-helix. J. Biol. Chem. 2000, 275, 18520–18526. [Google Scholar] [CrossRef]
- Tu, Y.; Woodson, J.; Ross, E.M. Binding of regulator of G protein signaling (RGS) proteins to phospholipid bilayers. Contribution of location and/or orientation to GTPase-activating protein activity. J. Biol. Chem. 2001, 276, 20160–20166. [Google Scholar] [CrossRef] [PubMed]
- Hope, M.J.; Cullis, P.R. The bilayer stability of inner monolayer lipids from the human erythrocyte. FEBS Lett. 1979, 107, 323–326. [Google Scholar] [CrossRef] [PubMed]
- Kirk, G.L.; Gruner, S.M.; Stein, D.L. A Thermodynamic Model of the Lamellar to Inverse Hexagonal Phase Transition of Lipid Membrane-Water Systems. Biochemistry 1984, 23, 1093–1102. [Google Scholar] [CrossRef]
- Matsuzaki, K.; Sugishita, K.; Ishibe, N.; Ueha, M.; Nakata, S.; Miyajima, K.; Epand, R.M. Relationship of membrane curvature to the formation of pores by magainin 2. Biochemistry 1998, 37, 11856–11863. [Google Scholar] [CrossRef] [PubMed]
- Liese, S.; Carlson, A. Membrane shape remodeling by protein crowding. Biophys. J. 2021, 120, 2482–2489. [Google Scholar] [CrossRef] [PubMed]
- Hirama, T.; Das, R.; Yang, Y.; Ferguson, C.; Won, A.; Yip, C.M.; Kay, J.G.; Grinstein, S.; Parton, R.G.; Fairn, G.D. Phosphatidylserine dictates the assembly and dynamics of caveolae in the plasma membrane. J. Biol. Chem. 2017, 292, 14292–14307. [Google Scholar] [CrossRef]
- Guyot, C.; Stieger, B. Interaction of bile salts with rat canalicular membrane vesicles: Evidence for bile salt resistant microdomains. J. Hepatol. 2011, 55, 1368–1376. [Google Scholar] [CrossRef]
- Allender, D.W.; Giang, H.; Schick, M. Model plasma mebrane exhibits a microemulsion in both leaves providing foundation for rafts. Biophys. J. 2020, 118, 1019–1031. [Google Scholar] [CrossRef]
- Huang, C.; Hepler, J.R.; Chen, L.T.; Gilman, A.G.; Anderson, R.G.; Mumby, S.M. Organization of G proteins and adenylyl cyclase at the plasma membrane. Mol. Biol. Cell 1997, 8, 2365–2378. [Google Scholar] [CrossRef] [Green Version]
- Sungkaworn, T.; Jobin, M.-L.; Burnecki, K.; Weron, A.; Lohse, M.J.; Calebiro, D. Single-molecule imaging reveals receptor-G protein interactions at cell surface hot spots. Nature 2017, 550, 543–547. [Google Scholar] [CrossRef] [PubMed]
- Taghon, G.J.; Rowe, J.B.; Kapolka, N.J.; Isom, D.G. Predictable cholesterol binding sites in GPCRs lack consensus motifs. Structure 2021, 29, 499–506. [Google Scholar] [CrossRef] [PubMed]
- Alemany, R.; Terés, S.; Baamonde, C.; Benet, M.; Vögler, O.; Escribá, P.V. 2-Hydroxyoleic acid: A new hypotensive molecule. Hypertension 2004, 43, 249–254. [Google Scholar] [CrossRef]
- Martínez, J.; Vögler, O.; Casas, J.; Barceló, F.; Alemany, R.; Prades, J.; Nagy, T.; Baamonde, C.; Kasprzyk, P.G.; Terés, S.; et al. Membrane structure modulation, protein kinase C alpha activation, and anticancer activity of minerval. Mol. Pharmacol. 2005, 67, 531–540. [Google Scholar] [CrossRef]
- Lladó, V.; Terés, S.; Higuera, M.; Alvarez, R.; Noguera-Salva, M.A.; Halver, J.E.; Escribá, P.V.; Busquets, X. Pivotal role of dihydrofolate reductase knockdown in the anticancer activity of 2-hydroxyoleic acid. Proc. Natl. Acad. Sci. USA 2009, 106, 13754–13758. [Google Scholar] [CrossRef] [PubMed]
- Lopez, D.H.; Fiol-deRoque, M.A.; Noguera-Salvà, M.A.; Terés, S.; Campana, F.; Piotto, S.; Castro, J.A.; Mohaibes, R.J.; Escribá, P.V.; Busquets, X. 2-Hydroxy arachidonic acid: A new non-steroidal anti-inflammatory drug. PLoS ONE 2013, 8, e72052. [Google Scholar] [CrossRef]
- Fiol-deRoque, M.A.; Gutierrez-Lanza, R.; Terés, S.; Torres, M.; Barceló, P.; Rial, R.V.; Verkhratsky, A.; Escribá, P.V.; Busquets, X.; Rodríguez, J.J. Cognitive recovery and restoration of cell proliferation in the dentate gyrus in the 5XFAD transgenic mice model of Alzheimer’s disease following 2-hydroxy-DHA treatment. Biogerontology 2013, 14, 763–775. [Google Scholar] [CrossRef]
- Torres, M.; Price, S.L.; Fiol-Deroque, M.A.; Marcilla-Etxenike, A.; Ahyayauch, H.; Barceló-Coblijn, G.; Terés, S.; Katsouri, L.; Ordinas, M.; López, D.J.; et al. Membrane lipid modifications and therapeutic effects mediated by hydroxydocosahexaenoic acid on Alzheimer’s disease. Biochim. Biophys. Acta 2014, 1838, 1680–1692. [Google Scholar] [CrossRef]
- Lladó, V.; López, D.J.; Ibarguren, M.; Alonso, M.; Soriano, J.B.; Escribá, P.V.; Busquets, X. Regulation of the cancer cell membrane lipid composition by NaCHOleate: Effects on cell signaling and therapeutical relevance in glioma. Biochim. Biophys. Acta 2014, 1838, 1619–1627. [Google Scholar] [CrossRef]
- Guardiola-Serrano, F.; Beteta-Göbel, R.; Rodríguez-Lorca, R.; Ibarguren, M.; López, D.J.; Terés, S.; Alvarez, R.; Alonso-Sande, M.; Busquets, X.; Escribá, P.V. The Novel Anticancer Drug Hydroxytriolein Inhibits Lung Cancer Cell Proliferation via a Protein Kinase Cα- and Extracellular Signal-Regulated Kinase 1/2-Dependent Mechanism. J. Pharmacol. Exp. Ther. 2015, 354, 213–224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torres, M.; Busquets, X.; Escribá, P.V. Brain Lipids in the Pathophysiology and Treatment of Alzheimer’s Disease. In Update on Dementia; Moretti, D.V., Ed.; InTech Open: London, UK, 2016; Volume 1, pp. 127–167. [Google Scholar]
- Alvarez, R.; Casas, J.; López, D.J.; Ibarguren, M.; Suari-Rivera, A.; Terés, S.; Guardiola-Serrano, F.; Lossos, A.; Busquets, X.; Kakhlon, O.; et al. Triacylglycerol mimetics regulate membrane interactions of glycogen branching enzyme: Implications for therapy. J. Lipid Res. 2017, 58, 1598–1612. [Google Scholar] [CrossRef] [PubMed]
- Vögler, O.; López-Bellan, A.; Alemany, R.; Tofé, S.; González, M.; Quevedo, J.; Pereg, V.; Barceló, F.; Escriba, P.V. Structure-effect relation of C18 long-chain fatty acids in the reduction of body weight in rats. Int. J. Obes. 2008, 32, 464–473. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, P.R.; Karunarathne, W.K.A.; Kalyanaraman, V.; Silvius, J.R.; Gautam, N. G-protein signaling leverages subunit-dependent membrane affinity to differentially control bg translocation to intracellular membranes. Proc. Natl. Acad. Sci. USA 2012, 109, E3568–E3577. [Google Scholar] [CrossRef] [Green Version]










| H2N-MASNNTASIAQARKLVEQLKMEANIDRIKVSKAAADLMAYCEAHAKEDPLL TPVPASENPFREKKFFCAIL-COOH |
| Proteins | Forward Oligonucleotide |
|---|---|
| 5′-ATCGAATTCATGGCCAGCAACAACACCGCCAGCATAGCACAAGCCAG-3′ | |
| Reverse Oligonucleotides | |
| wild type Gγ2 | 5′-CTCGCGGCCGCTTAAAGGATAGCACAGAA-3′ |
| GER- Gγ2 | 5′-CTCGCGGCCGCTTAAAGGATAGCAGAGAA-3′ |
| R62G Gγ2 | 5′-CTCGCGGCCGCTTAAAGGATAGCACAGAAAAACTTCTTCTCCCCAAA-3′ |
| K64G Gγ2 | 5′-CTCGCGGCCGCTTAAAGGATAGCACAGAAAAACTTCCCCTCCCTAAA-3′ |
| K65G Gγ2 | 5′-CTCGCGGCCGCTTAAAGGATAGCACAGAAAAACCCCTTCTCCCTAAA-3′ |
| R62G K64G Gγ2 | 5′-CTCGCGGCCGCTTAAAGGATAGCACAGAAAAACTTCCCCTCCCCAAA-3′ |
| R62G K65G Gγ2 | 5′-CTCGCGGCCGCTTAAAGGATAGCACAGAAAAACCCCTTCTCCCCAAA-3′ |
| K64G K65G Gγ2 | 5′-CTCGCGGCCGCTTAAAGGATAGCACAGAAAAACCCCCCCTCCCTAAA-3′ |
| R62G K64G K65G Gγ2 | 5′-CTCGCGGCCGCTTAAAGGATAGCACAGAAAAACCCCCCCTCCCCAAA-3′ |
| GER-R62G Gγ2 | 5′-CTCGCGGCCGCTTAAAGGATAGCAGAGAAAAACTTCTTCTCCCCAAA-3′ |
| GER-K64G Gγ2 | 5′-CTCGCGGCCGCTTAAAGGATAGCAGAGAAAAACTTCCCCTCCCTAAA-3′ |
| GER-K65G Gγ2 | 5′-CTCGCGGCCGCTTAAAGGATAGCAGAGAAAAACCCCTTCTCCCTAAA-3′ |
| GER-R62G K64G Gγ2 | 5′-CTCGCGGCCGCTTAAAGGATAGCAGAGAAAAACTTCCCCTCCCCAAA-3′ |
| GER-R62G K65G Gγ2 | 5′-CTCGCGGCCGCTTAAAGGATAGCAGAGAAAAACCCCTTCTCCCCAAA-3′ |
| GER-K64G K65G Gγ2 | 5′-CTCGCGGCCGCTTAAAGGATAGCAGAGAAAAACCCCCCCTCCCTAAA-3′ |
| GER-R62G K64G K65G Gγ2 | 5′-CTCGCGGCCGCTTAAAGGATAGCAGAGAAAAACCCCCCCTCCCCAAA-3′ |
| -MPVINIEDLTEKDKLKMEVDQLKKEVTLERMLVSKCCEEVRDYVEERSGEDPLVKGIPEDKNPFKELKGGCVIS | Gγ1 |
| ----MASN–NTASIAQARKLVEQLKMEANIDRIKVSKAAADLMAYCEAHAKEDPLLTPVPASENPFREKKFFCAIL | Gγ2 |
| MKGETPVN–STMSIGQARKMVEQLKIEASLCRIKVSKAAADLMTYCDAHACEDPLITPVPTSENPFREKKFFCALL | Gγ3 |
| KEGMSNN–STTSISQARKAVEQLKMEACMDRVKVSQAAADLLAYCEAHVREDPLIIPVPASENPFREKKFFCTIL | Gγ4 |
| ----MS----GSSSVAAMKKVVQQLRLEAGLNRVKVSQAAADLKQFCLQNAQHDPLLTGVSSSTNPFRPQKV-CSFL | Gγ5 |
| ----MS----ATNNIAQARKLVEQLRIEAGIERIKVSKAASDLMSYCEQHARNDPLLVGVPASENPFKDKKP-CIIL | Gγ6 |
| ----MS-N–NMAKIAEARKTVEQLKLEVNIDRMKVSQAAAELLAFCETHAKDDPLVTPVPAAENPFRDKRLFCVLL | Gγ7 |
| -------MAQDLSEKDLLKMEVEQLKKEVKNTRIPISKAGKEIKEYVEAQAGNDPFLKGIPEDKNPFKE-KGGCLIS | Gγ8 |
| ----MS----SGASASALQRLVEQLKLEAGVERIKVSQAAAELQQYCMQNACKDALLVGVPAGSNPFREPRS-CALL | Gγ9 |
| ---MPALHIEDLPEKEKLKMEVEQLRKEVKLQRQQVSKCSEEIKNYIEERSGEDPLVKGIPEDKNPFKE-KGSCVIS | Gγ10 |
| ----MSSKTASTNNIAQARRTVQQLRLEASIERIKVSKASADLMSYCEEHARSDPLLIGIPTSENPFKDKKT-CIIL | Gγ12 |
| ---------MEEWDVPQMKKEVESLKYQLAFQREMASKTIPELLKWIEDGIPKDPFLNPDLMKNNPWVE-KGKCTIL | Gγ13 |
| WT | PLLTPVPASENPFREKKFFCAIL | PLLTPVPASENPFREKKFFSAIL | ger- |
| K65 | PLLTPVPASENPFREKGFFCAIL | PLLTPVPASENPFREKGFFSAIL | ger-K65 |
| K64 | PLLTPVPASENPFREGKFFCAIL | PLLTPVPASENPFREGKFFSAIL | ger-K64 |
| R62 | PLLTPVPASENPFGEKKFFCAIL | PLLTPVPASENPFGEKKFFSAIL | ger-R62 |
| K64K65 | PLLTPVPASENPFREGGFFCAIL | PLLTPVPASENPFREGGFFSAIL | ger-K64K65 |
| R62K65 | PLLTPVPASENPFGEKGFFCAIL | PLLTPVPASENPFGEKGFFSAIL | ger-R62K65 |
| R62K64 | PLLTPVPASENPFGEGKFFCAIL | PLLTPVPASENPFGEGKFFSAIL | ger-R62K64 |
| R62K64K65 | PLLTPVPASENPFGEGGFFCAIL | PLLTPVPASENPFGEGGFFSAIL | ger-R62K64K65 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Álvarez, R.; Escribá, P.V. Structural Basis of the Interaction of the G Proteins, Gαi1, Gβ1γ2 and Gαi1β1γ2, with Membrane Microdomains and Their Relationship to Cell Localization and Activity. Biomedicines 2023, 11, 557. https://doi.org/10.3390/biomedicines11020557
Álvarez R, Escribá PV. Structural Basis of the Interaction of the G Proteins, Gαi1, Gβ1γ2 and Gαi1β1γ2, with Membrane Microdomains and Their Relationship to Cell Localization and Activity. Biomedicines. 2023; 11(2):557. https://doi.org/10.3390/biomedicines11020557
Chicago/Turabian StyleÁlvarez, Rafael, and Pablo V. Escribá. 2023. "Structural Basis of the Interaction of the G Proteins, Gαi1, Gβ1γ2 and Gαi1β1γ2, with Membrane Microdomains and Their Relationship to Cell Localization and Activity" Biomedicines 11, no. 2: 557. https://doi.org/10.3390/biomedicines11020557