Proteomic Profiling of Early Secreted Proteins in Response to Lipopolysaccharide-Induced Vascular Endothelial Cell EA.hy926 Injury
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemical and Reagents
2.2. Cell Type and Cell Culture
2.3. Determination of Cell Viability
2.4. Determination of Released Lactate Dehydrogenase (LDH) Activity
2.5. Determination of Apoptotic Cell Death
2.6. Actin Filaments Immunofluorescent Staining
2.7. Sample Preparation for Shotgun Proteomics
2.8. In-Solution Trypsin Digestion
2.9. Liquid Chromatography–Tandem Mass Spectrometry (LC-MS/MS)
2.10. Data Analysis
2.11. Statistical Analysis
3. Results
3.1. LPS-Induced Vascular Endothelial Cell Injury and Cell Death
3.2. LPS-Altered Actin Cytoskeletal Rearrangement and -Induced Apoptotic Cell Death
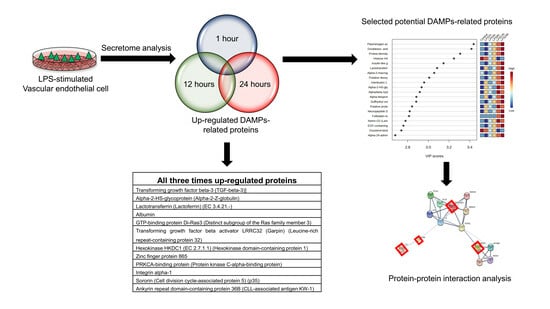
3.3. Identification of Vascular Endothelial Cell-Specific Protein–DAMPs in Response to LPS
3.4. Pairwise Comparisons of LPS Conditions
3.5. Identification of Candidate Proteins for Novel Early Sepsis Biomarkers
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Sepsis. Available online: https://www.who.int/news-room/fact-sheets/detail/sepsis (accessed on 20 August 2023).
- Rhodes, A.; Evans, L.E.; Alhazzani, W.; Levy, M.M.; Antonelli, M.; Ferrer, R.; Kumar, A.; Sevransky, J.E.; Sprung, C.L.; Nunnally, M.E.; et al. Surviving Sepsis Campaign: International Guidelines for Management of Sepsis and Septic Shock: 2016. Intensive Care Med. 2017, 43, 304–377. [Google Scholar] [CrossRef] [PubMed]
- Chun, K.; Syndergaard, C.; Damas, C.; Trubey, R.; Mukindaraj, A.; Qian, S.; Jin, X.; Breslow, S.; Niemz, A. Sepsis Pathogen Identification. J. Lab. Autom. 2015, 20, 539–561. [Google Scholar] [CrossRef] [PubMed]
- Dauphinee, S.M.; Karsan, A. Lipopolysaccharide signaling in endothelial cells. Lab. Investing. 2006, 86, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J.; Vincent, J.L.; Adhikari, N.K.; Machado, F.R.; Angus, D.C.; Calandra, T.; Jaton, K.; Giulieri, S.; Delaloye, J.; Opal, S.; et al. Sepsis: A roadmap for future research. Lancet. Infect. Dis. 2015, 15, 581–614. [Google Scholar] [CrossRef]
- Singer, M.; Deutschman, C.S.; Seymour, C.W.; Shankar-Hari, M.; Annane, D.; Bauer, M.; Bellomo, R.; Bernard, G.R.; Chiche, J.D.; Coopersmith, C.M.; et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016, 315, 801–810. [Google Scholar] [CrossRef]
- Barre, M.; Behnes, M.; Hamed, S.; Pauly, D.; Lepiorz, D.; Lang, S.; Akin, I.; Borggrefe, M.; Bertsch, T.; Hoffmann, U. Revisiting the prognostic value of monocyte chemotactic protein 1 and interleukin-6 in the sepsis-3 era. J. Crit. Care 2018, 43, 21–28. [Google Scholar] [CrossRef]
- Behnes, M.; Bertsch, T.; Lepiorz, D.; Lang, S.; Trinkmann, F.; Brueckmann, M.; Borggrefe, M.; Hoffmann, U. Diagnostic and prognostic utility of soluble CD 14 subtype (presepsin) for severe sepsis and septic shock during the first week of intensive care treatment. Crit. Care 2014, 18, 507. [Google Scholar] [CrossRef]
- Liu, S.; Wang, X.; She, F.; Zhang, W.; Liu, H.; Zhao, X. Effects of Neutrophil-to-Lymphocyte Ratio Combined with Interleukin-6 in Predicting 28-Day Mortality in Patients with Sepsis. Front. Immunol. 2021, 12, 639735. [Google Scholar] [CrossRef]
- Song, J.; Moon, S.; Park, D.W.; Cho, H.J.; Kim, J.Y.; Park, J.; Cha, J.H. Biomarker combination and SOFA score for the prediction of mortality in sepsis and septic shock: A prospective observational study according to the Sepsis-3 definitions. Medicine 2020, 99, e20495. [Google Scholar] [CrossRef]
- De Nooijer, A.H.; Pickkers, P.; Netea, M.G.; Kox, M. Inflammatory biomarkers to predict the prognosis of acute bacterial and viral infections. J. Crit. Care 2023, 78, 154360. [Google Scholar] [CrossRef]
- Zhou, M.; Aziz, M.; Wang, P. Damage-Associated Molecular Patterns as Double-Edged Swords in Sepsis. Antioxid. Redox Signal. 2021, 35, 1308–1323. [Google Scholar] [CrossRef] [PubMed]
- Itagaki, K.; Rica, I.; Konecna, B.; Kim, H.I.; Park, J.; Kaczmarek, E.; Hauser, C.J. Role of Mitochondria-Derived Danger Signals Released after Injury in Systemic Inflammation and Sepsis. Antioxid. Redox Signal. 2021, 35, 1273–1290. [Google Scholar] [CrossRef] [PubMed]
- Kongpol, K.; Nernpermpisooth, N.; Prompunt, E.; Kumphune, S. Endothelial-Cell-Derived Human Secretory Leukocyte Protease Inhibitor (SLPI) Protects Cardiomyocytes against Ischemia/Reperfusion Injury. Biomolecules 2019, 9, 678. [Google Scholar] [CrossRef]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
- Pang, Z.; Zhou, G.; Ewald, J.; Chang, L.; Hacariz, O.; Basu, N.; Xia, J. Using MetaboAnalyst 5.0 for LC–HRMS spectra processing, multi-omics integration and covariate adjustment of global metabolomics data. Nat. Protoc. 2022, 17, 1735–1761. [Google Scholar] [CrossRef]
- Zhao, T.; Wang, Z. GraphBio: A shiny web app to easily perform popular visualization analysis for omics data. Front. Genet. 2022, 13, 957317. [Google Scholar] [CrossRef]
- Mi, H.; Muruganujan, A.; Ebert, D.; Huang, X.; Thomas, P.D. PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019, 47, D419–D426. [Google Scholar] [CrossRef]
- Quoilin, C.; Mouithys-Mickalad, A.; Lécart, S.; Fontaine-Aupart, M.P.; Hoebeke, M. Evidence of oxidative stress and mitochondrial respiratory chain dysfunction in an in vitro model of sepsis-induced kidney injury. Biochim. Biophys. Acta (BBA)-Bioenerg. 2014, 1837, 1790–1800. [Google Scholar] [CrossRef]
- Li, W.; Deng, M.; Loughran, P.A.; Yang, M.; Lin, M.; Yang, C.; Gao, W.; Jin, S.; Li, S.; Cai, J.; et al. LPS Induces Active HMGB1 Release from Hepatocytes into Exosomes through the Coordinated Activities of TLR4 and Caspase-11/GSDMD Signaling. Front. Immunol. 2020, 11, 229. [Google Scholar] [CrossRef]
- Murao, A.; Aziz, M.; Wang, H.; Brenner, M.; Wang, P. Release mechanisms of major DAMPs. Apoptosis 2021, 26, 152–162. [Google Scholar] [CrossRef]
- Tang, D.; Chen, X.; Kang, R.; Kroemer, G. Ferroptosis: Molecular mechanisms and health implications. Cell Res. 2021, 31, 107–125. [Google Scholar] [CrossRef] [PubMed]
- Wen, Q.; Liu, J.; Kang, R.; Zhou, B.; Tang, D. The release and activity of HMGB1 in ferroptosis. Biochem. Biophys. Res. Commun. 2019, 510, 278–283. [Google Scholar] [CrossRef] [PubMed]
- Hossain, K.H.; Okamoto, T.; Usuda, H.; Jahan, I.; Niibayashi, T.; Wada, K. Differential Expression of Pro-inflammatory and Pro-coagulant Genes in Endothelial Cells Induced by Porphyromonas gingivalis Lipopolysaccharide, Escherichia coli Lipopolysaccharide, and Zymosan. Shimane J. Med. Sci. 2020, 37, 123–132. [Google Scholar] [CrossRef]
- Kwon, O.K.; Lee, W.; Kim, S.J.; Lee, Y.M.; Lee, J.Y.; Kim, J.Y.; Bae, J.S.; Lee, S. In-depth proteomics approach of secretome to identify novel biomarker for sepsis in LPS-stimulated endothelial cells. Electrophoresis 2015, 36, 2851–2858. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Timilsena, Y.P.; Blanch, E.; Adhikari, B. Lactoferrin: Structure, function, denaturation and digestion. Crit. Rev. Food Sci. Nutr. 2019, 59, 580–596. [Google Scholar] [CrossRef]
- Tong, Y.; Ku, X.; Wu, C.; Liu, J.; Yang, C.; Tang, W.; Yan, W.; Tang, J. Data-independent acquisition-based quantitative proteomic analysis reveals differences in host immune response of peripheral blood mononuclear cells to sepsis. Scand. J. Immunol. 2019, 89, e12748. [Google Scholar] [CrossRef]
- Lin, Y.H.; Zhu, J.; Meijer, S.; Franc, V.; Heck, A.J.R. Glycoproteogenomics: A frequent gene polymorphism affects the glycosylation pattern of the human serum fetuin/alpha-2-HS-glycoprotein. Mol. Cell. Proteom. MCP 2019, 18, 1479–1490. [Google Scholar] [CrossRef]
- Wang, H.; Sama, A.E. Anti-inflammatory role of fetuin-A in injury and infection. Curr. Mol. Med. 2012, 12, 625–633. [Google Scholar] [CrossRef]
- Degirmencioglu, H.; Ozer Bekmez, B.; Derme, T.; Oncel, M.Y.; Canpolat, F.E.; Tayman, C. Presepsin and fetuin-A dyad for the diagnosis of proven sepsis in preterm neonates. BMC Infect. Dis. 2019, 19, 695. [Google Scholar] [CrossRef] [PubMed]
- Turgman, O.; Schinkel, M.; Wiersinga, W.J. Host response biomarkers for sepsis in the emergency room. Crit. Care 2023, 27, 97. [Google Scholar] [CrossRef] [PubMed]
- Pilar-Orive, F.J.A.I.; Azkargorta, M.; Elortza, F.; Garcia-Obregon, S. A three-protein panel to support the diagnosis of sepsis in children. J. Clin. Med. 2022, 11, 1563. [Google Scholar] [CrossRef] [PubMed]
- Baranska, P.; Jerczynska, H.; Pawlowska, Z.; Koziolkiewicz, W.; Cierniewski, C.S. Expression of Integrins and Adhesive Properties of Human Endothelial Cell Line EA.hy 926. Cancer Genom. Proteom. 2005, 2, 265–269. [Google Scholar]
- Bauer, J.; Margolis, M.; Schreiner, C.; Edgell, C.J.; Azizkhan, J.; Lazarowski, E.; Juliano, R.L. In vitro model of angiogenesis using a human endothelium-derived permanent cell line: Contributions of induced gene expression, G-proteins, and integrins. J. Cell. Physiol. 1992, 153, 437–449. [Google Scholar] [CrossRef]
- Siqueiros-Cendón, T.; Arévalo-Gallegos, S.; Iglesias-Figueroa, B.F.; García-Montoya, I.A.; Salazar-Martínez, J.; Rascón-Cruz, Q. Immunomodulatory effects of lactoferrin. Acta Pharmacol. Sin. 2014, 35, 557–566. [Google Scholar] [CrossRef]
- Kan, C.; Yang, J.; Fan, H.; Dai, Y.; Wang, X.; Chen, R.; Liu, J.; Meng, X.; Wang, W.; Li, G.; et al. Fetuin-A is an immunomodulator and a potential therapeutic option in BMP4-dependent heterotopic ossification and associated bone mass loss. Bone Res. 2022, 10, 62. [Google Scholar] [CrossRef]





| Protein Names | f. Value | p. Value | −log10p | FDR |
|---|---|---|---|---|
| Transforming growth factor beta-3 (TGF-beta-3) | 26.734 | 7.57 × 10−9 | 8.121 | 1.53 × 10−5 |
| Alpha-2-HS-glycoprotein (alpha-2-Z-globulin) | 21.088 | 1.00 × 10−7 | 6.9986 | 5.08 × 10−5 |
| Lactotransferrin (lactoferrin) (EC 3.4.21.-) | 19.989 | 1.75 × 10−7 | 6.7575 | 7.08 × 10−5 |
| Albumin | 14.497 | 3.89 × 10−6 | 5.4101 | 0.000988 |
| GTP-binding protein Di-Ras3 (distinct subgroup of the Ras family member 3) | 14.25 | 4.54 × 10−6 | 5.3428 | 0.001022 |
| Transforming growth factor beta activator LRRC32 (garpin) (leucine-rich repeat domain-containing protein 32) | 12.867 | 1.11 × 10−5 | 4.955 | 0.002043 |
| Hexokinase HKDC1 (EC 2.7.1.1) (hexokinase domain-containing protein 1) | 11.876 | 2.17 × 10−5 | 4.6632 | 0.003384 |
| Zinc finger protein 865 | 10.661 | 5.15 × 10−5 | 4.2884 | 0.007448 |
| PRKCA-binding protein (protein kinase C-alpha-binding protein) | 9.5197 | 0.000121 | 3.9175 | 0.013742 |
| Integrin alpha-1 | 7.6929 | 0.000524 | 3.281 | 0.037017 |
| Sororin (cell division cycle-associated protein 5) (p35) | 7.6665 | 0.000535 | 3.2713 | 0.037017 |
| Ankyrin repeat domain-containing protein 36B (CLL-associated antigen KW-1) | 7.3272 | 0.000714 | 3.1466 | 0.041303 |
| Pathway ID | Pathway Description | False Discovery Rate |
|---|---|---|
| GO:0080134 | regulation of response to stress | 4.40 × 10−9 |
| GO:0051707 | response to other organism | 7.76 × 10−8 |
| GO:0009617 | response to bacterium | 8.73 × 10−8 |
| GO:0031347 | regulation of defense response | 8.73 × 10−8 |
| GO:0048583 | regulation of response to stimuli | 1.37 × 10−7 |
| GO:0032496 | response to lipopolysaccharide | 5.06 × 10−6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Songjang, W.; Paiyabhroma, N.; Jumroon, N.; Jiraviriyakul, A.; Nernpermpisooth, N.; Seenak, P.; Kumphune, S.; Thaisakun, S.; Phaonakrop, N.; Roytrakul, S.; et al. Proteomic Profiling of Early Secreted Proteins in Response to Lipopolysaccharide-Induced Vascular Endothelial Cell EA.hy926 Injury. Biomedicines 2023, 11, 3065. https://doi.org/10.3390/biomedicines11113065
Songjang W, Paiyabhroma N, Jumroon N, Jiraviriyakul A, Nernpermpisooth N, Seenak P, Kumphune S, Thaisakun S, Phaonakrop N, Roytrakul S, et al. Proteomic Profiling of Early Secreted Proteins in Response to Lipopolysaccharide-Induced Vascular Endothelial Cell EA.hy926 Injury. Biomedicines. 2023; 11(11):3065. https://doi.org/10.3390/biomedicines11113065
Chicago/Turabian StyleSongjang, Worawat, Nitchawat Paiyabhroma, Noppadon Jumroon, Arunya Jiraviriyakul, Nitirut Nernpermpisooth, Porrnthanate Seenak, Sarawut Kumphune, Siriwan Thaisakun, Narumon Phaonakrop, Sittiruk Roytrakul, and et al. 2023. "Proteomic Profiling of Early Secreted Proteins in Response to Lipopolysaccharide-Induced Vascular Endothelial Cell EA.hy926 Injury" Biomedicines 11, no. 11: 3065. https://doi.org/10.3390/biomedicines11113065