Molecular Cloning, Heterologous Expression, Purification, and Evaluation of Protein–Ligand Interactions of CYP51 of Candida krusei Azole-Resistant Fungal Strain
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Modeling of C. krusei CYP51 3D Structure
2.3. Molecular Cloning and Expression of C. krusei CYP51
- C. krusei_fusF: ttaggaggtcatatgcatcatcatcatcatcattccgtcatcaaggcaat;
- C. krusei_fusR: taagcttcgtcatcagttcttttgtcttctctc.
2.4. Purification of C. krusei CYP51
2.5. Reconstitution Activity Assay
2.6. Ligand Screening
2.7. Spectrophotometric Titration
2.8. Mass Spectrometric Analysis
2.9. Obtaining of Chlorotopsenthiasterol Sulfate D (S-232-Cl) and Granulatoside A (Ch-4)
3. Results
3.1. Comparative Analysis of CYP51 of Clinically Relevant Fungi of the Genus Candida
3.2. Molecular Cloning, Expression, and Purification of C. krusei CYP51
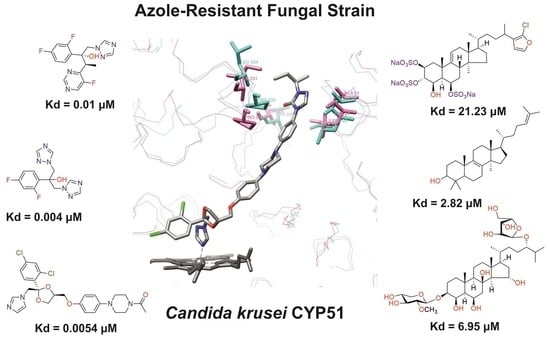
3.3. Ligand-Binding Properties of CYP51 C. krusei
3.3.1. Interaction of C. krusei CYP51 with Azole Antifungals
3.3.2. Interaction of C. krusei CYP51 with Steroid Derivatives
4. Discussion
- -
- Obtain highly purified proteins of pathogenic organisms, especially resistant strains, that are targets for the action of drug compounds;
- -
- Investigate the interaction of molecular drug targets with known drugs;
- -
- Search for potential ligands of molecular targets that have a chemical scaffold different from that of existing drugs.
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sahu, S.R.; Bose, S.; Singh, M.; Kumari, P.; Dutta, A.; Utkalaja, B.G.; Patel, S.K.; Acharya, N. Vaccines against candidiasis: Status, challenges and emerging opportunity. Front. Cell. Infect. Microbiol. 2022, 12, 1002406. [Google Scholar] [CrossRef]
- Jamiu, A.T.; Albertyn, J.; Sebolai, O.M.; Pohl, C.H. Update on Candida krusei, a potential multidrug-resistant pathogen. Med. Mycol. 2021, 59, 14–30. [Google Scholar] [CrossRef]
- Bassetti, M.; Peghin, M.; Timsit, J.F. The current treatment landscape: Candidiasis. J. Antimicrob. Chemother. 2016, 71 (Suppl. S2), ii13–ii22. [Google Scholar] [CrossRef]
- Bouopda Tamo, S.P. Candida Infections: Clinical Features, Diagnosis and Treatment. Infect. Dis. Clin. Microbiol. 2020, 2, 91–102. [Google Scholar] [CrossRef]
- Gomez-Gaviria, M.; Ramirez-Sotelo, U.; Mora-Montes, H.M. Non-albicans Candida Species: Immune Response, Evasion Mechanisms, and New Plant-Derived Alternative Therapies. J. Fungi 2022, 9, 11. [Google Scholar] [CrossRef]
- Guinea, J. Global trends in the distribution of Candida species causing candidemia. Clin. Microbiol. Infect. 2014, 20 (Suppl. S6), 5–10. [Google Scholar] [CrossRef] [PubMed]
- Pappas, P.G.; Lionakis, M.S.; Arendrup, M.C.; Ostrosky-Zeichner, L.; Kullberg, B.J. Invasive candidiasis. Nat. Rev. Dis. Primers 2018, 4, 18026. [Google Scholar] [CrossRef]
- Pfaller, M.A.; Diekema, D.J.; Turnidge, J.D.; Castanheira, M.; Jones, R.N. Twenty Years of the SENTRY Antifungal Surveillance Program: Results for Candida Species from 1997–2016. Open. Forum Infect. Dis. 2019, 6 (Suppl. S1), S79–S94. [Google Scholar] [CrossRef]
- Esfahani, A.; Omran, A.N.; Salehi, Z.; Shams-Ghahfarokhi, M.; Ghane, M.; Eybpoosh, S.; Razzaghi-Abyaneh, M. Molecular epidemiology, antifungal susceptibility, and ERG11 gene mutation of Candida species isolated from vulvovaginal candidiasis: Comparison between recurrent and non-recurrent infections. Microb. Pathog. 2022, 170, 105696. [Google Scholar] [CrossRef]
- Arendrup, M.C.; Patterson, T.F. Multidrug-Resistant Candida: Epidemiology, Molecular Mechanisms, and Treatment. J. Infect. Dis. 2017, 216 (Suppl. S3), S445–S451. [Google Scholar] [CrossRef]
- von Lilienfeld-Toal, M.; Wagener, J.; Einsele, H.; Cornely, O.A.; Kurzai, O. Invasive Fungal Infection. Dtsch. Arztebl. Int. 2019, 116, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Kumaraswamy, M.; Coady, A.; Szubin, R.; Martin, T.C.; Palsson, B.; Nizet, V.; Monk, J.M. Comprehensive whole genome sequencing with hybrid assembly of multi-drug resistant Candida albicans isolate causing cerebral abscess. Curr. Res. Microb. Sci. 2023, 4, 100180. [Google Scholar] [CrossRef] [PubMed]
- Cortegiani, A.; Misseri, G.; Fasciana, T.; Giammanco, A.; Giarratano, A.; Chowdhary, A. Epidemiology, clinical characteristics, resistance, and treatment of infections by Candida auris. J. Intensive Care 2018, 6, 69. [Google Scholar] [CrossRef]
- Yang, B.; Rao, R. Emerging Pathogens of the Candida Species. In Candida Albicans, 1st ed.; Sandai, D., Ed.; Intech Open Science: London, UK, 2019; pp. 3–18. [Google Scholar] [CrossRef]
- Olver, W.J.; Scott, F.; Shankland, G.S. Successful treatment of Candida krusei fungemia with amphotericin B and caspofungin. Med. Mycol. 2006, 44, 655–657. [Google Scholar] [CrossRef] [PubMed]
- Schilling, A.; Seibold, M.; Mansmann, V.; Gleissner, B. Successfully treated Candida krusei infection of the lumbar spine with combined caspofungin/posaconazole therapy. Med. Mycol. 2008, 46, 79–83. [Google Scholar] [CrossRef]
- Katragkou, A.; McCarthy, M.; Meletiadis, J.; Hussain, K.; Moradi, P.W.; Strauss, G.E.; Myint, K.L.; Zaw, M.H.; Kovanda, L.L.; Petraitiene, R.; et al. In vitro combination therapy with isavuconazole against Candida spp. Med. Mycol. 2017, 55, 859–868. [Google Scholar] [CrossRef]
- Chapman, S.W.; Sullivan, D.C.; Cleary, J.D. In search of the holy grail of antifungal therapy. Trans. Am. Clin. Climatol. Assoc. 2008, 119, 197–215. [Google Scholar]
- Shafiei, M.; Peyton, L.; Hashemzadeh, M.; Foroumadi, A. History of the development of antifungal azoles: A review on structures, SAR, and mechanism of action. Bioorg. Chem. 2020, 104, 104240. [Google Scholar] [CrossRef]
- Becher, R.; Wirsel, S.G. Fungal cytochrome P450 sterol 14alpha-demethylase (CYP51) and azole resistance in plant and human pathogens. Appl. Microbiol. Biotechnol. 2012, 95, 825–840. [Google Scholar] [CrossRef]
- Monk, B.C.; Sagatova, A.A.; Hosseini, P.; Ruma, Y.N.; Wilson, R.K.; Keniya, M.V. Fungal Lanosterol 14alpha-demethylase: A target for next-generation antifungal design. Biochim. Biophys. Acta-Proteins Proteom. 2020, 1868, 140206. [Google Scholar] [CrossRef]
- Hassan, S.T.; Berchova-Bimova, K.; Petras, J. Plumbagin, a Plant-Derived Compound, Exhibits Antifungal Combinatory Effect with Amphotericin B against Candida albicans Clinical Isolates and Anti-hepatitis C Virus Activity. Phytother. Res. PTR 2016, 30, 1487–1492. [Google Scholar] [CrossRef] [PubMed]
- Bezerra, C.F.; Rocha, J.E.; Nascimento Silva, M.K.D.; de Freitas, T.S.; de Sousa, A.K.; Dos Santos, A.T.L.; da Cruz, R.P.; Ferreira, M.H.; da Silva, J.C.P.; Machado, A.J.T.; et al. Analysis by UPLC-MS-QTOF and antifungal activity of guava (Psidium guajava L.). Food Chem. Toxicol. 2018, 119, 122–132. [Google Scholar] [CrossRef] [PubMed]
- Souza-Moreira, T.M.; Severi, J.A.; Rodrigues, E.R.; de Paula, M.I.; Freitas, J.A.; Vilegas, W.; Pietro, R. Flavonoids from Plinia cauliflora (Mart.) Kausel (Myrtaceae) with antifungal activity. Nat. Prod. Res. 2019, 33, 2579–2582. [Google Scholar] [CrossRef]
- Fukuoka, T.; Johnston, D.A.; Winslow, C.A.; de Groot, M.J.; Burt, C.; Hitchcock, C.A.; Filler, S.G. Genetic basis for differential activities of fluconazole and voriconazole against Candida krusei. Antimicrob. Agents Chemother. 2003, 47, 1213–1219. [Google Scholar] [CrossRef] [PubMed]
- Morio, F.; Loge, C.; Besse, B.; Hennequin, C.; Le Pape, P. Screening for amino acid substitutions in the Candida albicans Erg11 protein of azole-susceptible and azole-resistant clinical isolates: New substitutions and a review of the literature. Diagn. Microbiol. Infect. Dis. 2010, 66, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Flowers, S.A.; Colon, B.; Whaley, S.G.; Schuler, M.A.; Rogers, P.D. Contribution of clinically derived mutations in ERG11 to azole resistance in Candida albicans. Antimicrob. Agents Chemother. 2015, 59, 450–460. [Google Scholar] [CrossRef]
- Warrilow, A.G.; Nishimoto, A.T.; Parker, J.E.; Price, C.L.; Flowers, S.A.; Kelly, D.E.; Rogers, P.D.; Kelly, S.L. The Evolution of Azole Resistance in Candida albicans Sterol 14alpha-Demethylase (CYP51) through Incremental Amino Acid Substitutions. Antimicrob. Agents Chemother. 2019, 63, e02586-18. [Google Scholar] [CrossRef]
- Grossman, N.T.; Pham, C.D.; Cleveland, A.A.; Lockhart, S.R. Molecular mechanisms of fluconazole resistance in Candida parapsilosis isolates from a U.S. surveillance system. Antimicrob. Agents Chemother. 2015, 59, 1030–1037. [Google Scholar] [CrossRef]
- Tan, J.; Zhang, J.; Chen, W.; Sun, Y.; Wan, Z.; Li, R.; Liu, W. The A395T mutation in ERG11 gene confers fluconazole resistance in Candida tropicalis causing candidemia. Mycopathologia 2015, 179, 213–218. [Google Scholar] [CrossRef]
- Ksiezopolska, E.; Gabaldon, T. Evolutionary Emergence of Drug Resistance in Candida Opportunistic Pathogens. Genes 2018, 9, 461. [Google Scholar] [CrossRef]
- Mirdita, M.; Schutze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making protein folding accessible to all. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Omura, T.; Sato, R. The Carbon Monoxide-Binding Pigment of Liver Microsomes. I. Evidence for Its Hemoprotein Nature. J. Biol. Chem. 1964, 239, 2370–2378. [Google Scholar] [CrossRef] [PubMed]
- Guengerich, F.P.; Martin, M.V.; Sohl, C.D.; Cheng, Q. Measurement of cytochrome P450 and NADPH-cytochrome P450 reductase. Nat. Protoc. 2009, 4, 1245–1251. [Google Scholar] [CrossRef] [PubMed]
- Kaluzhskiy, L.; Ershov, P.; Yablokov, E.; Shkel, T.; Grabovec, I.; Mezentsev, Y.; Gnedenko, O.; Usanov, S.; Shabunya, P.; Fatykhava, S.; et al. Human Lanosterol 14-Alpha Demethylase (CYP51A1) Is a Putative Target for Natural Flavonoid Luteolin 7,3′-Disulfate. Molecules 2021, 26, 2237. [Google Scholar] [CrossRef]
- Strushkevich, N.; Usanov, S.A.; Park, H.W. Structural basis of human CYP51 inhibition by antifungal azoles. J. Mol. Biol. 2010, 397, 1067–1078. [Google Scholar] [CrossRef]
- Ouellet, H.; Guan, S.; Johnston, J.B.; Chow, E.D.; Kells, P.M.; Burlingame, A.L.; Cox, J.S.; Podust, L.M.; de Montellano, P.R. Mycobacterium tuberculosis CYP125A1, a steroid C27 monooxygenase that detoxifies intracellularly generated cholest-4-en-3-one. Mol. Microbiol. 2010, 77, 730–742. [Google Scholar] [CrossRef]
- Tabakmakher, K.M.; Makarieva, T.N.; Denisenko, V.A.; Popov, R.S.; Dmitrenok, P.S.; Dyshlovoy, S.A.; Grebnev, B.B.; Bokemeyer, C.; von Amsberg, G.; Cuong, N.X. New Trisulfated Steroids from the Vietnamese Marine Sponge Halichondria vansoesti and Their PSA Expression and Glucose Uptake Inhibitory Activities. Mar. Drugs 2019, 17, 445. [Google Scholar] [CrossRef]
- Ivanchina, N.V.; Kicha, A.A.; Malyarenko, T.V.; Ermolaeva, S.D.; Yurchenko, E.A.; Pislyagin, E.A.; Van Minh, C.; Dmitrenok, P.S. Granulatosides D, E and other polar steroid compounds from the starfish Choriaster granulatus. Their immunomodulatory activity and cytotoxicity. Nat. Prod. Res. 2019, 33, 2623–2630. [Google Scholar] [CrossRef]
- Xiang, M.J.; Liu, J.Y.; Ni, P.H.; Wang, S.; Shi, C.; Wei, B.; Ni, Y.X.; Ge, H.L. Erg11 mutations associated with azole resistance in clinical isolates of Candida albicans. FEMS Yeast Res. 2013, 13, 386–393. [Google Scholar] [CrossRef]
- Wu, Y.; Gao, N.; Li, C.; Gao, J.; Ying, C. A newly identified amino acid substitution T123I in the 14alpha-demethylase (Erg11p) of Candida albicans confers azole resistance. FEMS Yeast Res. 2017, 17, fox012. [Google Scholar] [CrossRef] [PubMed]
- Perea, S.; Lopez-Ribot, J.L.; Kirkpatrick, W.R.; McAtee, R.K.; Santillan, R.A.; Martinez, M.; Calabrese, D.; Sanglard, D.; Patterson, T.F. Prevalence of molecular mechanisms of resistance to azole antifungal agents in Candida albicans strains displaying high-level fluconazole resistance isolated from human immunodeficiency virus-infected patients. Antimicrob. Agents Chemother. 2001, 45, 2676–2684. [Google Scholar] [CrossRef] [PubMed]
- Sanglard, D.; Ischer, F.; Koymans, L.; Bille, J. Amino acid substitutions in the cytochrome P-450 lanosterol 14alpha-demethylase (CYP51A1) from azole-resistant Candida albicans clinical isolates contribute to resistance to azole antifungal agents. Antimicrob. Agents Chemother. 1998, 42, 241–253. [Google Scholar] [CrossRef] [PubMed]
- Silva, D.B.; Rodrigues, L.M.; Almeida, A.A.; Oliveira, K.M.; Grisolia, A.B. Novel point mutations in the ERG11 gene in clinical isolates of azole resistant Candida species. Mem. Inst. Oswaldo Cruz 2016, 111, 192–199. [Google Scholar] [CrossRef] [PubMed]
- Favre, B.; Didmon, M.; Ryder, N.S. Multiple amino acid substitutions in lanosterol 14alpha-demethylase contribute to azole resistance in Candida albicans. Microbiology 1999, 145, 2715–2725. [Google Scholar] [CrossRef] [PubMed]
- Manavathu, E.K.; Kallakuri, S.; Arganoza, M.T.; Vazquez, J.A. Amino acid variations of cytochrome P-450 lanosterol 14 alpha-demethylase (CYP51A1) from fluconazoleresistant clinical isolates of Candida albicans. Rev. Iberoam. Micol. 1999, 16, 198–203. [Google Scholar]
- Chau, A.S.; Mendrick, C.A.; Sabatelli, F.J.; Loebenberg, D.; McNicholas, P.M. Application of real-time quantitative PCR to molecular analysis of Candida albicans strains exhibiting reduced susceptibility to azoles. Antimicrob. Agents Chemother. 2004, 48, 2124–2131. [Google Scholar] [CrossRef]
- White, T.C. The presence of an R467K amino acid substitution and loss of allelic variation correlate with an azole-resistant lanosterol 14alpha demethylase in Candida albicans. Antimicrob. Agents Chemother. 1997, 41, 1488–1494. [Google Scholar] [CrossRef]
- Wang, H.; Kong, F.; Sorrell, T.C.; Wang, B.; McNicholas, P.; Pantarat, N.; Ellis, D.; Xiao, M.; Widmer, F.; Chen, S.C. Rapid detection of ERG11 gene mutations in clinical Candida albicans isolates with reduced susceptibility to fluconazole by rolling circle amplification and DNA sequencing. BMC Microbiol. 2009, 9, 167. [Google Scholar] [CrossRef]
- Shkel, T.V.; Vasilevskaya, A.V.; Gilep, A.A.; Usanov, S.A.; Chernovetsky, M.A.; Lukyanenko, I.G. Molecular cloning, expression, purification and ligand-binding properties of CYP51 from Candida albicans. Dokl. Akad. Nauk 2012, 56, 64–71. (In Russian) [Google Scholar]
- Kaluzhskiy, L.A.; Shkel, T.V.; Ivanchina, N.V.; Kicha, A.A.; Grabovec, I.P.; Gilep, A.A.; Strushkevich, N.V.; Chernovetsky, M.A.; Medvedev, A.E.; Usanov, S.A.; et al. Structural analogues of lanosterol from marine organisms of the class Asteroidea as potential inhibitors of human and Candida albicans lanosterol 14α-demethylases. Nat. Product Comm. 2017, 12, 1843–1846. [Google Scholar] [CrossRef]
- Lewis, D.F.V. (Ed.) Introduction. In Guide to Cytochromes P450 Structure and Function, 2nd ed.; Taylor & Francis: London, UK; New York, NY, USA, 2001; pp. 2–18. [Google Scholar]
- Firooz, A.; Nafisi, S.; Maibach, H.I. Novel drug delivery strategies for improving econazole antifungal action. Int. J. Pharm. 2015, 495, 599–607. [Google Scholar] [CrossRef] [PubMed]
- Kale, P.; Johnson, L.B. Second-generation azole antifungal agents. Drugs Today 2005, 41, 91–105. [Google Scholar] [CrossRef] [PubMed]
- Ivanchina, N.V.; Kicha, A.A.; Stonik, V.A. Steroid glycosides from marine organisms. Steroids 2011, 76, 425–454. [Google Scholar] [CrossRef]
- Kaluzhskiy, L.; Tsybruk, T.; Yablokov, E.; Gnedenko, O.; Zelepuga, E.; Kicha, A.; Kozlovskaya, E.; Ivanchina, N.; Gilep, A.; Ivanov, A. SPR Biosensor Based High-Throughput Screening of Low Molecular Weight Compounds for Interaction with Candida krusei CYP51. Biomed. Chem. Res. Methods 2023, 6, e00183. [Google Scholar] [CrossRef]
- Parker, J.E.; Warrilow, A.G.; Price, C.L.; Mullins, J.G.; Kelly, D.E.; Kelly, S.L. Resistance to antifungals that target CYP51. J. Chem. Biol. 2014, 7, 143–161. [Google Scholar] [CrossRef]
- He, X.; Zhao, M.; Chen, J.; Wu, R.; Zhang, J.; Cui, R.; Jiang, Y.; Chen, J.; Cao, X.; Xing, Y.; et al. Overexpression of Both ERG11 and ABC2 Genes Might Be Responsible for Itraconazole Resistance in Clinical Isolates of Candida krusei. PLoS ONE 2015, 10, e0136185. [Google Scholar] [CrossRef]
- Cowen, L.E.; Sanglard, D.; Howard, S.J.; Rogers, P.D.; Perlin, D.S. Mechanisms of Antifungal Drug Resistance. Cold Spring Harb. Perspect. Med. 2014, 5, a019752. [Google Scholar] [CrossRef]
- Whaley, S.G.; Berkow, E.L.; Rybak, J.M.; Nishimoto, A.T.; Barker, K.S.; Rogers, P.D. Azole Antifungal Resistance in Candida albicans and Emerging Non-albicans Candida Species. Front. Microbiol. 2016, 7, 2173. [Google Scholar] [CrossRef]
- Hargrove, T.Y.; Garvey, E.P.; Hoekstra, W.J.; Yates, C.M.; Wawrzak, Z.; Rachakonda, G.; Villalta, F.; Lepesheva, G.I. Crystal Structure of the New Investigational Drug Candidate VT-1598 in Complex with Aspergillus fumigatus Sterol 14alpha-Demethylase Provides Insights into Its Broad-Spectrum Antifungal Activity. Antimicrob. Agents Chemother. 2017, 61, e00570-17. [Google Scholar] [CrossRef]






| Compound | Structure | Spectrum | Kd (µM), ΔAmax, Type of Spectral Response |
|---|---|---|---|
| Ketoconazole |  |  | Kd = 0.0054 ± 0.0026 Titration range: 0.02–2.26 µM ΔAmax = 0.070 type II |
| Bifonazole |  |  | Kd = 0.003 ± 0.001 Titration range: 0.02–1.22 µM ΔAmax = 0.050 type II |
| Clotrimazole |  |  | Kd = 0.0013 ± 0.0008 Titration range: 0.02–1.14 µM ΔAmax = 0.051 type II |
| Econazole |  |  | Kd = 0.0013 ± 0.0004 Titration range: 0.004–0.83 µM ΔAmax = 0.058 type II |
| Fluconazole |  |  | Kd = 0.004 ± 0.001 Titration range: 0.02–1.08 µM ΔAmax = 0.031 type II |
| Miconazole |  |  | Kd = 0.002 ± 0.0009 Titration range: 0.02–1.48 µM ΔAmax = 0.048 type II |
| Voriconazole |  |  | Kd = 0.01 ± 0.006 Titration range: 0.02–3.12 µM ΔAmax = 0.023 type II |
| Compound | Structure | Spectrum | Kd, µM; ΔAmax; Type of Spectral Response |
|---|---|---|---|
| Lanosterol |  |  | Kd = 2.82 ± 0.18 Titration range: 0.1–30.9 µM ΔAmax = 0.031 type I |
| 73c (14,17-etheno-3-hydroxy-16α-nitroestra-1,3,5(10)-trien-17β-yl acetate) |  |  | Kd = 10.7 ± 1.5 Titration range: 0.1–44.7 µM ΔAmax = 0.0069 type I |
| 99 (3β,20-dihydroxy-24-hydroxyiminocholest-5,22-diene) |  |  | Kd = 1.02 ± 0.14 Titration range: 0.1–10.7 µM ΔAmax = 0.0047 type I |
| S-232-Cl (chlorotopsenthiasterol sulfate D, sponge Halichondria vansoesti) |  |  | Kd = 21.23 ± 2.83 Titration range: 0.1–140.5 µM ΔAmax = 0.0034 type I |
| Ch-4 (granulatoside A, starfish Choriaster granulatus) |  |  | Kd = 6.95 ± 0.80 Titration range: 0.1–95.5 µM ΔAmax = 0.0099 type I |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsybruk, T.V.; Kaluzhskiy, L.A.; Mezentsev, Y.V.; Makarieva, T.N.; Tabakmaher, K.M.; Ivanchina, N.V.; Dmitrenok, P.S.; Baranovsky, A.V.; Gilep, A.A.; Ivanov, A.S. Molecular Cloning, Heterologous Expression, Purification, and Evaluation of Protein–Ligand Interactions of CYP51 of Candida krusei Azole-Resistant Fungal Strain. Biomedicines 2023, 11, 2873. https://doi.org/10.3390/biomedicines11112873
Tsybruk TV, Kaluzhskiy LA, Mezentsev YV, Makarieva TN, Tabakmaher KM, Ivanchina NV, Dmitrenok PS, Baranovsky AV, Gilep AA, Ivanov AS. Molecular Cloning, Heterologous Expression, Purification, and Evaluation of Protein–Ligand Interactions of CYP51 of Candida krusei Azole-Resistant Fungal Strain. Biomedicines. 2023; 11(11):2873. https://doi.org/10.3390/biomedicines11112873
Chicago/Turabian StyleTsybruk, Tatsiana V., Leonid A. Kaluzhskiy, Yuri V. Mezentsev, Tatyana N. Makarieva, Kseniya M. Tabakmaher, Natalia V. Ivanchina, Pavel S. Dmitrenok, Alexander V. Baranovsky, Andrei A. Gilep, and Alexis S. Ivanov. 2023. "Molecular Cloning, Heterologous Expression, Purification, and Evaluation of Protein–Ligand Interactions of CYP51 of Candida krusei Azole-Resistant Fungal Strain" Biomedicines 11, no. 11: 2873. https://doi.org/10.3390/biomedicines11112873