Advancing Nanoscale Science: Synthesis and Bioprinting of Zeolitic Imidazole Framework-8 for Enhanced Anti-Infectious Therapeutic Efficacies
Abstract
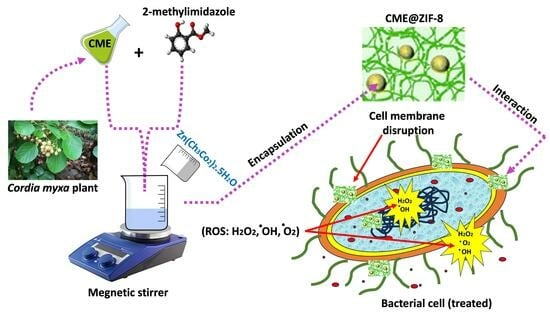
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Plant Extract Preparation
2.3. Synthesis of ZIF-8
2.4. CME@ZIF-8 Synthesis
2.5. Characterization
2.6. Antimicrobial Tests
2.6.1. Disc Diffusion Method
2.6.2. Minimum Inhibitory Concentration
3. Results and Discussion
3.1. Physico-Chemical Characterizations of Prepared MOFs
3.2. Antibacterial Activity of MOFs
3.2.1. Disc Diffusion Method
3.2.2. Minimum Inhibitory Concentration (MIC)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Duan, S.; Duan, S.; Zhao, X.; Su, Z.; Wang, C.; Lin, Y. Layer-by-Layer Decorated Nanoscale ZIF-8 with High Curcumin Loading Effectively Inactivates Gram-Negative and Gram-Positive Bacteria. ACS Appl. Bio Mater. 2020, 3, 3673–3680. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Peng, F.; Wang, D. MOFs and MOF-Derived Materials for Antibacterial Application. J. Funct. Biomater. 2022, 13, 215. [Google Scholar] [CrossRef] [PubMed]
- Matthiessen, L.; Bergström, R.; Dustdar, S.; Meulien, P.; Draghia-Akli, R. Increased Momentum in Antimicrobial Resistance Research. Lancet 2016, 388, 865. [Google Scholar] [CrossRef]
- Ahanger, A.M.; Kumar, S.; Arya, A.; Suryavanshi, A.; Kain, D. Vandana Synthesis and Encapsulation of Ajuga Parviflora Extract with Zeolitic Imidazolate Framework-8 and Their Therapeutic Action against G+and G-Drug-Resistant Bacteria. ACS Omega 2022, 7, 1671–1681. [Google Scholar] [CrossRef] [PubMed]
- Rubinstein, E.; Kollef, M.H.; Nathwani, D. Pneumonia Caused by Methicillin-Resistant Staphylococcus Aureus. Clin. Infect. Dis. 2008, 46, S378–S385. [Google Scholar] [CrossRef]
- Park, S.H. Third-Generation Cephalosporin Resistance in Gram-Negative Bacteria in the Community: A Growing Public Health Concern. Korean J. Intern. Med. 2014, 29, 27–30. [Google Scholar] [CrossRef]
- Iyer, M. ICMR Report; Times of India: Mumbai, India, 2021. [Google Scholar]
- Li, C.; Sun, F. Graphene-Assisted Sensor for Rapid Detection of Antibiotic Resistance in Escherichia Coli. Front. Chem. 2021, 9, 696906. [Google Scholar] [CrossRef]
- Rasheed, M.U.; Thajuddin, N.; Ahamed, P.; Teklemariam, Z.; Jamil, K. Resistência Microbiana a Drogas Em Linhagens de Escherichia Coli Isoladas de Fontes Alimentares. Rev. Inst. Med. Trop. Sao Paulo 2014, 56, 341–346. [Google Scholar] [CrossRef]
- Wang, L.; Hu, C.; Shao, L. The Antimicrobial Activity of Nanoparticles: Present Situation and Prospects for the Future. Int. J. Nanomed. 2017, 12, 1227–1249. [Google Scholar] [CrossRef]
- Hetta, H.F.; Ramadan, Y.N.; Al-Harbi, A.I.; Ahmed, E.A.; Battah, B.; Abd Ellah, N.H.; Zanetti, S.; Donadu, M.G. Nanotechnology as a Promising Approach to Combat Multidrug Resistant Bacteria: A Comprehensive Review and Future Perspectives. Biomedicines 2023, 11, 413. [Google Scholar] [CrossRef]
- Ahmed, S.A.; Bagchi, D.; Katouah, H.A.; Hasan, M.N.; Altass, H.M.; Pal, S.K. Enhanced Water Stability and Photoresponsivity in Metal-Organic Framework (MOF): A Potential Tool to Combat Drug-Resistant Bacteria. Sci. Rep. 2019, 9, 19372. [Google Scholar] [CrossRef]
- Lai, D.; Zhou, A.; Tan, B.K.; Tang, Y.; Sarah Hamzah, S.; Zhang, Z.; Lin, S.; Hu, J. Preparation and Photodynamic Bactericidal Effects of Curcumin-β-Cyclodextrin Complex. Food Chem. 2021, 361, 130117. [Google Scholar] [CrossRef] [PubMed]
- El-Massry, A.; Ibrahim, O.; Abdalla, M.; Osman, I.; Mahmoud, S. Safety and Indicative Effectiveness of Porcine Corneal Lenticular Implants in Patients with Advanced Keratoconus and Post Lasik Ectasia: A Retrospective Clinical Study. Clin. Ophthalmol. 2021, 15, 3165–3171. [Google Scholar] [CrossRef] [PubMed]
- Batool, S.; Hasan, M.; Dilshad, M.; Zafar, A.; Tariq, T.; Wu, Z.; Chen, R.; Gul Hassan, S.; Munawar, T.; Iqbal, F.; et al. Green Synthesis of Cordia Myxa Incubated ZnO, Fe2O3, and CO3O4 Nanoparticle: Characterization, and Their Response as Biological and Photocatalytic Agent. Adv. Powder Technol. 2022, 33, 103780. [Google Scholar] [CrossRef]
- Murthy, H.; Iqbal, M.; Chavez, J.C.; Kharfan-Dabaja, M.A. Cytokine Release Syndrome: Current Perspectives. Immunotargets Ther. 2019, 8, 43–52. [Google Scholar] [CrossRef]
- Maleki, A.; Shahbazi, M.A.; Alinezhad, V.; Santos, H.A. The Progress and Prospect of Zeolitic Imidazolate Frameworks in Cancer Therapy, Antibacterial Activity, and Biomineralization. Adv. Healthc. Mater. 2020, 9, e2000248. [Google Scholar] [CrossRef]
- Menezes, J.E.S.A.; dos Santos, H.S.; Ferreira, M.K.A.; Magalhães, F.E.A.; da Silva, D.S.; Bandeira, P.N.; Saraiva, G.D.; Pessoa, O.D.L.; Ricardo, N.M.P.S.; Cruz, B.G.; et al. Preparation, Structural and Spectroscopic Characterization of Chitosan Membranes Containing Allantoin. J. Mol. Struct. 2020, 1199, 126968. [Google Scholar] [CrossRef]
- Kargar, H.; Ghahramaninezhad, M.; Shahrak, M.N.; Balula, S.S. An Effective Magnetic Catalyst for Oxidative Desulfurization of Model and Real Fuels: Fe3O4/ZIF-8/TiO2. Microporous Mesoporous Mater. 2021, 317, 110992. [Google Scholar] [CrossRef]
- Kang, L.; Smith, S.; Wang, C. Stabilization of Surface-Bound Antibodies for ELISA Based on a Reversable Zeolitic Imidazolate Framework-8 Coating. J. Colloid Interface Sci. 2021, 588, 101–109. [Google Scholar] [CrossRef]
- Li, H.; Gao, X.; Jia, C.; Chen, W.; Liu, B.; Yang, L.; Sun, C.; Chen, G. Enrichment of Hydrogen from a Hydrogen/Propylene Gas Mixture Using Zif-8/Water-Glycol Slurry. Energies 2018, 11, 1890. [Google Scholar] [CrossRef]
- Jameh, A.A.; Mohammadi, T.; Bakhtiari, O.; Mahdyarfar, M. Synthesis and Modification of Zeolitic Imidazolate Framework (ZIF-8) Nanoparticles as Highly Efficient Adsorbent for H2S and CO2 Removal from Natural Gas. J. Environ. Chem. Eng. 2019, 7, 103058. [Google Scholar] [CrossRef]
- Ghaffar, I.; Imran, M.; Perveen, S.; Kanwal, T.; Saifullah, S.; Bertino, M.F.; Ehrhardt, C.J.; Yadavalli, V.K.; Shah, M.R. Synthesis of Chitosan Coated Metal Organic Frameworks (MOFs) for Increasing Vancomycin Bactericidal Potentials against Resistant S. Aureus Strain. Mater. Sci. Eng. C 2019, 105, 110111. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, N.; Pandey, S.K.; Mehta, J.; Bhardwaj, S.K.; Kim, K.H.; Deep, A. Bioactive Nano-Metal-Organic Frameworks as Antimicrobials against Gram-Positive and Gram-Negative Bacteria. Toxicol. Res. 2018, 7, 931–941. [Google Scholar] [CrossRef] [PubMed]
- Aryanejad, S.; Bagherzade, G.; Moudi, M. Design and Development of Novel Co-MOF Nanostructures as an Excellent Catalyst for Alcohol Oxidation and Henry Reaction, with a Potential Antibacterial Activity. Appl. Organomet. Chem. 2019, 33, e4820. [Google Scholar] [CrossRef]
- Abd El Salam, H.M.; Nassar, H.N.; Khidr, A.S.A.; Zaki, T. Antimicrobial Activities of Green Synthesized Ag Nanoparticles @ Ni-MOF Nanosheets. J. Inorg. Organomet. Polym. Mater. 2018, 28, 2791–2798. [Google Scholar] [CrossRef]
- Kaur, H.; Mohanta, G.C.; Gupta, V.; Kukkar, D.; Tyagi, S. Synthesis and Characterization of ZIF-8 Nanoparticles for Controlled Release of 6-Mercaptopurine Drug. J. Drug Deliv. Sci. Technol. 2017, 41, 106–112. [Google Scholar] [CrossRef]
- Saif, M.S.; Zafar, A.; Waqas, M.; Hassan, S.G.; ul Haq, A.; Tariq, T.; Batool, S.; Dilshad, M.; Hasan, M.; Shu, X. Phyto-Reflexive Zinc Oxide Nano-Flowers Synthesis: An Advanced Photocatalytic Degradation and Infectious Therapy. J. Mater. Res. Technol. 2021, 13, 2375–2391. [Google Scholar] [CrossRef]
- Geethalakshmi, R.; Sarada, D.V.L. Evaluation of Antimicrobial and Antioxidant Activity of Essential Oil of Trianthema decandra L. J. Pharm. Res. 2013, 6, 101–106. [Google Scholar] [CrossRef]
- Narra, N.; Kaki, S.S.; Prasad, R.B.N.; Misra, S.; Dhevendar, K.; Kontham, V.; Korlipara, P.V. Synthesis and Evaluation of Anti-Oxidant and Cytotoxic Activities of Novel 10-Undecenoic Acid Methyl Ester Based Lipoconjugates of Phenolic Acids. Beilstein J. Org. Chem. 2017, 13, 26–32. [Google Scholar] [CrossRef]
- Shaaban, M.T.; Ghaly, M.F.; Fahmi, S.M. Antibacterial Activities of Hexadecanoic Acid Methyl Ester and Green-Synthesized Silver Nanoparticles against Multidrug-Resistant Bacteria. J. Basic Microbiol. 2021, 61, 557–568. [Google Scholar] [CrossRef]
- Ganesan, T.; Subban, M.; Christopher Leslee, D.B.; Kuppannan, S.B.; Seedevi, P. Structural Characterization of N-Hexadecanoic Acid from the Leaves of Ipomoea Eriocarpa and Its Antioxidant and Antibacterial Activities. Biomass Convers. Biorefinery 2022. [Google Scholar] [CrossRef]
- Siswadi, S.; Saragih, G.S. Phytochemical Analysis of Bioactive Compounds in Ethanolic Extract of Sterculia Quadrifida R.Br. In Proceedings of the AIP Conference Proceedings; American Institute of Physics Inc.: College Park, MD, USA, 2021; Volume 2353. [Google Scholar]
- Rahman, M.M.; Ahmad, S.H.; Mohamed, M.T.M.; Ab Rahman, M.Z. Antimicrobial Compounds from Leaf Extracts of Jatropha Curcas, Psidium Guajava, and Andrographis Paniculata. Sci. World J. 2014, 2014, 635240. [Google Scholar] [CrossRef] [PubMed]
- Mazumder, K.; Nabila, A.; Aktar, A.; Farahnaky, A. Bioactive Variability and in Vitro and in Vivo Antioxidant Activity of Unprocessed and Processed Flour of Nine Cultivars of Australian Lupin Species: A Comprehensive Substantiation. Antioxidants 2020, 9, 282. [Google Scholar] [CrossRef] [PubMed]
- Desbois, A.P.; Smith, V.J. Antibacterial Free Fatty Acids: Activities, Mechanisms of Action and Biotechnological Potential. Appl. Microbiol. Biotechnol. 2010, 85, 1629–1642. [Google Scholar] [CrossRef]
- Adebayo, I.A.; Arsad, H.; Samian, M.R. Methyl Elaidate: A Major Compound of Potential Anticancer Extract of Moringa Oleifera Seeds Binds with Bax and MDM2 (P53 Inhibitor) in Silico. Pharmacogn. Mag. 2018, 14, S554–S557. [Google Scholar] [CrossRef]
- Li, Y.; Hu, H.-B.; Zheng, X.-D.; Zhu, J.-H.; Liu, L.-P. Composition and Antimicrobial Activity of Essential Oil from the Aerial Part of Artemisia Annua. J. Med. Plants Res. 2011, 5, 3629–3633. [Google Scholar]
- Yabalak, E.; Erdoğan Eliuz, E.A.; Nazlı, M.D. Evaluation of Citrus Reticulata Essential Oil: Chemical Composition and Antibacterial Effectiveness Incorporated Gelatin on E. coli and S. aureus. Int. J. Environ. Health Res. 2022, 32, 1261–1270. [Google Scholar] [CrossRef]
- Nakaziba, R.; Amanya, S.B.; Sesaazi, C.D.; Byarugaba, F.; Ogwal-Okeng, J.; Alele, P.E. Antimicrobial Bioactivity and GC-MS Analysis of Different Extracts of Corchorus olitorius L. Leaves. Sci. World J. 2022, 2022, 3382302. [Google Scholar] [CrossRef]
- Hatami, S.; Mohamadi Sani, A.; Yavarmanesh, M. Chemical Composition and Antibacterial Activity of Organic Extra Virgin Olive Oil from Iran. Nutr. Food Sci. 2016, 46, 388–395. [Google Scholar] [CrossRef]
- Zheng, C.J.; Yoo, J.S.; Lee, T.G.; Cho, H.Y.; Kim, Y.H.; Kim, W.G. Fatty Acid Synthesis Is a Target for Antibacterial Activity of Unsaturated Fatty Acids. FEBS Lett. 2005, 579, 5157–5162. [Google Scholar] [CrossRef]
- Elzein Hagr, T.; Abd elmageed, M.; Eldeen Abdlrazig, S.H.; Salim Holi, A.; Hamadnalla, H.M.Y. GS-MS Study, Antimicrobial and Antioxidant Activity of Fixed Oil from Ximenia Americana L. Seeds. Open Access J. Chem. 2020, 4, 8–13. [Google Scholar] [CrossRef]
- Swantara, M.D.; Rita, W.S.; Suartha, N.; Agustina, K.K. Anticancer Activities of Toxic Isolate of Xestospongia Testudinaria Sponge. Vet. World 2019, 12, 1434–1440. [Google Scholar] [CrossRef] [PubMed]
- Ghareeb, M.A.; Hamdi, S.A.H.; Fol, M.F.; Ibrahim, A.M. Chemical Characterization, Antibacterial, Antibiofilm, and Antioxidant Activities of the Methanolic Extract of Paratapes Undulatus Clams (Born, 1778). J. Appl. Pharm. Sci. 2022, 12, 219–228. [Google Scholar] [CrossRef]
- Karimi, E.; Ze Jaafar, H.; Ghasemzadeh, A.; Ebrahimi, M. Fatty Acid Composition, Antioxidant and Antibacterial Properties of the Microwave Aqueous Extract of Three Varieties of Labisia Pumila Benth. Biol. Res. 2015, 48, 9. [Google Scholar] [CrossRef]
- Boulet, J.C.; Ducasse, M.A.; Cheynier, V. Ultraviolet Spectroscopy Study of Phenolic Substances and Other Major Compounds in Red Wines: Relationship between Astringency and the Concentration of Phenolic Substances. Aust. J. Grape Wine Res. 2017, 23, 193–199. [Google Scholar] [CrossRef]
- Wu, C.; Liu, Q.; Chen, R.; Liu, J.; Zhang, H.; Li, R.; Takahashi, K.; Liu, P.; Wang, J. Fabrication of ZIF-8@SiO2 Micro/Nano Hierarchical Superhydrophobic Surface on AZ31 Magnesium Alloy with Impressive Corrosion Resistance and Abrasion Resistance. ACS Appl. Mater. Interfaces 2017, 9, 11106–11115. [Google Scholar] [CrossRef]
- Schejn, A.; Balan, L.; Falk, V.; Aranda, L.; Medjahdi, G.; Schneider, R. Controlling ZIF-8 Nano- and Microcrystal Formation and Reactivity through Zinc Salt Variations. CrystEngComm 2014, 16, 4493–4500. [Google Scholar] [CrossRef]
- Nguyen, L.T.L.; Le, K.K.A.; Phan, N.T.S. A Zeolite Imidazolate Framework ZIF-8 Catalyst for Friedel-Crafts Acylation. Cuihua Xuebao/Chin. J. Catal. 2012, 33, 688–696. [Google Scholar] [CrossRef]
- Sasaki, K.; Okue, T.; Shu, Y.; Miyake, K.; Uchida, Y.; Nishiyama, N. Thin ZIF-8 Nanosheets Synthesized in Hydrophilic TRAPs. Dalton Trans. 2021, 50, 10394–10399. [Google Scholar] [CrossRef]
- García-Palacín, M.; Martínez, J.I.; Paseta, L.; Deacon, A.; Johnson, T.; Malankowska, M.; Téllez, C.; Coronas, J. Sized-Controlled ZIF-8 Nanoparticle Synthesis from Recycled Mother Liquors: Environmental Impact Assessment. ACS Sustain. Chem. Eng. 2020, 8, 2973–2980. [Google Scholar] [CrossRef]
- Ge, X.; Wong, R.; Anisa, A.; Ma, S. Recent Development of Metal-Organic Framework Nanocomposites for Biomedical Applications. Biomaterials 2022, 281, 121322. [Google Scholar] [CrossRef] [PubMed]
- Nordin, N.A.H.M.; Ismail, A.F.; Mustafa, A.; Murali, R.S.; Matsuura, T. The Impact of ZIF-8 Particle Size and Heat Treatment on CO2/CH4 Separation Using Asymmetric Mixed Matrix Membrane. RSC Adv. 2014, 4, 52530–52541. [Google Scholar] [CrossRef]
- Ali, A.; Saeed, S.; Hussain, R.; Afzal, G.; Siddique, A.B.; Parveen, G.; Hasan, M.; Caprioli, G. Synthesis and Characterization of Silica, Silver-Silica, and Zinc Oxide-Silica Nanoparticles for Evaluation of Blood Biochemistry, Oxidative Stress, and Hepatotoxicity in Albino Rats. ACS Omega 2023, 8, 20900–20911. [Google Scholar] [CrossRef] [PubMed]
- Binaeian, E.; Maleki, S.; Motaghedi, N.; Arjmandi, M. Study on the Performance of Cd2+ Sorption Using Dimethylethylenediamine-Modified Zinc-Based MOF (ZIF-8-Mmen): Optimization of the Process by RSM Technique. Sep. Sci. Technol. 2020, 55, 2713–2728. [Google Scholar] [CrossRef]
- Tian, J.; An, M.; Zhao, X.; Wang, Y.; Hasan, M. Advances in Fluorescent Sensing Carbon Dots: An Account of Food Analysis. ACS Omega 2023, 8, 9031–9039. [Google Scholar] [CrossRef]
- Meroueh, S.O.; Bencze, K.Z.; Hesek, D.; Lee, M.; Fisher, J.F.; Stemmler, T.L.; Mobashery, S. Three-dimensional structure of the bacterial cell wall peptidoglycan. Proc. Natl. Acad. Sci. USA 2006, 103, 4404–4409. [Google Scholar] [CrossRef]
- Quijia, C.R.; Alves, R.C.; Hanck-Silva, G.; Galvão Frem, R.C.; Arroyos, G.; Chorilli, M. Metal-Organic Frameworks for Diagnosis and Therapy of Infectious Diseases. Crit. Rev. Microbiol. 2022, 48, 161–196. [Google Scholar] [CrossRef]








| No | RT (m) | Area | Compound Name | MF | Mol. Wt. g/mol | Properties |
|---|---|---|---|---|---|---|
| 1 | 15.115 | 4651 | 3,8-dimethylundecane | C12H26 | 170.33 | Antibacterial [29] |
| 2 | 23.817 | 9778 | Undecanoic acid,10-methyl-, methyl ester | C13H26O2 | 214.34 | Anticancer [30] |
| 3 | 24.965 | 279,571 | Hexadecanoic acid, methyl ester | C17H34O2 | 270.5 | Antibacterial [31] |
| 4 | 25.805 | 16,632 | n-Hexadecanoic acid | C16H32O2 | 256.42 | Antimicrobial [32], antioxidant, and anti-inflammatory [33] |
| 5 | 28.213 | 24,517 | Methyl (9E,12E)-9,12-octadecadienoate | C19H34O2 | 294.5 | Antibacterial [34], Antioxidant [35] |
| 6 | 28.334 | 127,970 | Oleic acid, methyl ester | C19H36O2 | 296.5 | Antibacterial [36] |
| 7 | 28.461 | 48,494 | Methyl (9E)-9-octadecenoate | C19H36O2 | 296.5 | Anticancer [37], antibacterial, and antioxidant [38,39] |
| 8 | 28.845 | 205,937 | Stearic acid, methyl ester | C19H38O2 | 298.5 | Antibacterial [40] |
| 9 | 29.235 | 32,457 | 9-Octadecenal, (Z)- | C18H34O2 | 266.5 | Antibacterial and antifungal [41] |
| 10 | 29.357 | 17,037 | cis,cis-Linoleic acid | C18H32O2 | 280.45 | Antimicrobial, antioxidant [42] |
| No | RT (m) | Area | Compound Name | MF | Mol. Wt. g/mol | Properties |
|---|---|---|---|---|---|---|
| 1 | 21.722 | 70,816 | Methyl 8-(2-octyl cyclopropyl) octanoate | C20H38O2 | 310.5 | Antibacterial and antioxidant [43] |
| 2 | 23.817 | 698,232 | Hexadecanoic acid, methyl ester | C17H34O2 | 270.5 | Antibacterial [31] |
| 3 | 27.648 | 5650 | 1,2-Cyclohexanedimethanol | C8H16O2 | 144.21 | -- |
| 4 | 27.879 | 5229 | 1,E-11,Z-13-Octadecatriene | C16H32O2 | 256.42 | Anticancer [44] |
| 5 | 28.210 | 67,605 | Methyl (9E,12E)-9,12-octadecadienoate | C19H34O2 | 294.5 | Antibacterial, antioxidant [35] |
| 6 | 28.333 | 365,536 | Oleic acid, methyl ester | C19H36O2 | 296.5 | Antibacterial, antimicrobial [36] |
| 7 | 28.459 | 128,105 | 7-Hexadecenoic acid, methyl ester, (Z)- | C17H32O2 | 268.4 | Antibacterial and antioxidant [45] |
| 8 | 28.842 | 526,375 | Stearic acid, methyl ester | C19H38O2 | 298.5 | Antibacterial [46] |
| 9 | 29.017 | 14,779 | (1-Methyl-1-propylpentyl) benzene | C15H24 | 204.35 | -- |
| 10 | 29.266 | 18,651 | Nopyl acetate | C13H20O2 | 208.30 | -- |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saif, M.S.; Hasan, M.; Zafar, A.; Ahmed, M.M.; Tariq, T.; Waqas, M.; Hussain, R.; Zafar, A.; Xue, H.; Shu, X. Advancing Nanoscale Science: Synthesis and Bioprinting of Zeolitic Imidazole Framework-8 for Enhanced Anti-Infectious Therapeutic Efficacies. Biomedicines 2023, 11, 2832. https://doi.org/10.3390/biomedicines11102832
Saif MS, Hasan M, Zafar A, Ahmed MM, Tariq T, Waqas M, Hussain R, Zafar A, Xue H, Shu X. Advancing Nanoscale Science: Synthesis and Bioprinting of Zeolitic Imidazole Framework-8 for Enhanced Anti-Infectious Therapeutic Efficacies. Biomedicines. 2023; 11(10):2832. https://doi.org/10.3390/biomedicines11102832
Chicago/Turabian StyleSaif, Muhammad Saqib, Murtaza Hasan, Ayesha Zafar, Muhammad Mahmood Ahmed, Tuba Tariq, Muhammad Waqas, Riaz Hussain, Amna Zafar, Huang Xue, and Xugang Shu. 2023. "Advancing Nanoscale Science: Synthesis and Bioprinting of Zeolitic Imidazole Framework-8 for Enhanced Anti-Infectious Therapeutic Efficacies" Biomedicines 11, no. 10: 2832. https://doi.org/10.3390/biomedicines11102832